Abstract
Mutations in leucine-repeat rich kinase-2 (LRRK2) are the most common known cause of late-onset Parkinson's disease. In this study, a novel system to purify active recombinant LRRK2 expressed in mammalian cells was generated. This recombinant enzyme was used to characterize the specificity of LRRK2 and identify small compounds that can inhibit the kinase activity. Recombinant LRRK2 was shown to autophosphorylate and phosphorylate MBP and a peptide (LRRKtide) corresponding to the T688 site in moesin. A series of well-characterized kinase peptide substrates was not modified by LRRK2 demonstrating remarkable specificity. G2019S, the most common disease-causing mutation in LRRK2, increased kinase activity more dramatically than previously appreciated (∼10-fold). Several small molecules sharing a basic indolocarbazole structure (Gö6976, K252a and staurosporine) where identified as potent inhibitors of LRRK2 kinase activity. These findings provide important insights and tools to study the mechanisms of LRRK2 pathobiology, and could lead to therapeutic applications.
Keywords: inhibitors, kinase, LRRK2, Parkinson's disease, PARK8, substrates
Introduction
Parkinson's disease (PD) is the most common neurodegenerative movement disorder. The principal pathologies of PD are the loss of dopaminergic neurons in the substantia nigra pars compacta in addition to the presence of intracytoplasmic inclusions known as Lewy bodies and Lewy neurites in some of the remaining dopaminergic neurons [1; 2]. The mechanism(s) and cellular insults leading to the demise of neurons in PD are under investigation; however oxidative stress, protein misfolding, and mitochondrial dysfunction have been implicated as factors in the disease process [3-5].
The discovery of specific genes that can be causal of PD has provided important new insights into the cellular pathways that are involved in the pathobiology of PD [6-9]. Studies in the past few years have revealed that missense autosomal dominant mutations in the leucine-rich repeat kinase 2 (LRRK2) gene (also known as PARK8) are collectively the single most commonly known cause of late-onset PD [10-12]. However, the G2019S mutation is the most common, and it is responsible for 2–8% of hereditary PD, and 1-2 % of sporadic PD cases [12-18]. In North African Arabs and Ashkenazi Jews, the G2019S has been reported to be causal of 20-40% of PD cases [19-21].
LRRK2 is a large 2527 amino acid protein with several discrete domains that include many leucine-rich repeats, a Ras (renin-angiotensin system) of complex (ROC) domain, which belongs to the Ras GTPase superfamily, and a kinase domain towards the C-terminal end [12; 22; 23]. In vitro, LRRK2 functions as a Ser/Thr kinase that can undergo autophosphorylation, and can phosphorylate the generic kinase substrate myelin basic protein (MBP) [24-26]. It was recently shown that T558 in moesin can be phosphorylated by LRRK2 [26]. The G2019S mutation appears to affect the activation loop of the kinase domain and in some in vitro studies it results in 2-3 fold increase in kinase activity [26-28] which may lead to neurotoxicity [25; 28; 29]. Nevertheless, there is still limited information on the specificity of LRRK2, and inhibitors for this kinase have not been reported. In this study we describe a novel system to purify active recombinant LRRK2. This recombinant enzyme was used to characterize the specificity of LRRK2 and identify the first potent molecular inhibitors of LRRK2 kinase activity.
Materials and Methods
Materials
Goat anti-glutathione-S-transferase (GST) polyclonal antibody was purchased from Amersham Biosciences (Piscataway, NJ). 1182 is a rabbit polyclonal antibody raised against a recombinant His-tagged LRRK2 protein fragment corresponding to amino acid residues 841-960. The shuttling vector pCR8/GW/TOPO and the mammalian expression GST-tagged vector pDEST27 were purchased from Invitrogen (Carlsbad, CA). Bovine MBP and the synthetic peptides Kemptide (LRRASLG), caesin kinase 2 substrate (RRRADDSD), MBP fragment 104-118 (GKGRGLSLSRFSWGA), and [Ser25]-PKC fragment 19-31 (RFARKGSLRQKNV) were purchased from Sigma-Aldrich (St. Louis, MO). The synthetic peptides MBP fragment 4-14 (KRPSQRSKYL), MBP fragment 94-102 (APRTPGGRR), MARCKS-derived peptide (KKRFSFKKSFKL), and PKC-delta peptide substrate (RFAVRDMRQTVAVGVIKAVDKK) were purchased from Calbiochem/EMD Biosciences (Gibbstown, NJ). LRRKtide (RLGRDKYKTLRQIRQ) was synthesized and purified on reverse phase HPLC by the W.M Keck Biotechnology Resource Center at Yale University, New Haven, CT. The following kinase inhibitors were purchased from Calbiochem/EMD Biosciences: IC261, 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), Gö6976, H89, K-252a, K-252b, kenpaullone, KN-62, LY294002, ML-7, olomoucine, PD98059, Raf1 kinase inhibitor I, rapamycin, roscovitine, SB203580, staurosporine, U0126, wortmannin, Y-27632.
Cell Culture
Human embryonic kidney 293T cells (293T) were cultured in Dulbecco's modified medium (DMEM) high glucose (4.5gm/L) supplemented with 10 % fetal bovine serum (FBS), 100U/ml penicillin, 100U/ml streptomycin, and 2mM L-glutamine.
LRRK2 Expression Constructs
The full-length human LRRK2 cDNA was amplified by PCR using Taq polymerase AccuPrime SuperMix (Invitrogen) and cloned by topoisomerase reaction into the shuttling vector pCR8/GW/TOPO. To generate the cDNA encoding the G2019S mutation, the LRRK2 cDNA fragment spanning the AvrII and NcoI restriction sites in LRRK2 was amplified by PCR and cloned by topoisomerase reaction into the vector pCR4-TOPO (Invitrogen). The mutation corresponding to the G2019S amino acid substitution was generated using the QuickChange® Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA). A LRRK2 AvrII/NcoI cDNA fragment containing the LRRK2 “triple kinase-dead” (TKD) mutant [28] was amplified by PCR using a plasmid kindly provided by Dr. Mark Cookson and cloned in the vector pCR4-TOPO. In this TKD mutant, the amino acid responsible for ATP binding (K1906A), the active site aspartate (D1994A), and the Mg2+ binding site (D2017A) ware mutated. The AvrII/NcoI DNA fragments containing either the G2019S or TDK mutant were reintroduced into full-length LRRK2 by subcloning with these restriction enzymes. The sequence of the plasmids was verified by DNA sequencing using primers that span the whole cDNA as a service offered by the DNA Sequencing Facility of the University of Pennsylvania.
WT and mutants full-length LRRK2 cDNAs were introduced into the pDEST27 vector by recombinase reaction using LR Clonase II enzyme (Invitrogen) to generate a plasmid expressing N-terminal GST-tagged protein.
Western Blotting Analysis
Proteins were resolved on SDS-polyacrylamide gels by SDS-PAGE and electrophoretically transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Hercules, Ca) in buffer containing 190 mM glycine, 25 mM Tris-base and 10 % methanol. Membranes were blocked with a 5% powdered skimmed milk solution dissolved in Tris buffered saline (50 mM Tris, pH 7.6, 150 mM NaCl), incubated with primary antibody followed with an anti-goat antibody conjugated to horse radish peroxidase, developed with Western Lightning Chemiluminescence Reagents (PerkinElmer Life Sciences (Boston, MA) and exposed onto X-Omat Blue XB-1 films (Kodak, Rochester, NY).
In-vitro LRRK2 Kinase assays
293T cells were transiently transfected with pDEST27/LRRK2 expression plasmid using calcium phosphate precipitation method buffered with N, N-bis(2-hydroxyethyl)-2-amino-ethanesulfonic acid (BES) [30]. 48-72 hours after transfection, cells were washed and harvested with PBS, and resuspended in lysis buffer (25nM Tris pH 7.4, 5nM EDTA, 10mM beta-glycerol phosphate, 1mM NaVO4, 1 % Triton X-100, 0.5% glycerol with protease inhibitor cocktail) at 4°C. Cell debris were removed by sedimentation at 13,000×g for 15 min, and supernatants were precleared by incubation with sepharose beads that were removed by sedimentation. Supernatants were incubated with glutathione-sepharose beads (GE Healthcare) for 3 hrs at 4°C. Beads were extensively washed with lysis buffer (5 times) and wash buffer (25 mM Hepes, pH. 7.4, 1mM DTT, 10 mM β-glycerophosphate)(5 times) and eluted with wash buffer with 20mM glutathione. The kinase reactions were conducted at 25°C by incubating purified GST-LRRK2 in 25 μL of kinase buffer (25 mM Hepes, pH 7.4, 1mM DTT, 10 mM β-glycerophosphate, 10 mM MnCl2, 1 μM ATP, 5uCi [γ-32P]ATP) with 0.04 mg/ml MBP or 0.04 mg/ml synthetic peptide. For autophosphorylation or phosphorylation of MBP, reactions were stopped with the addition of sodium dodecyl sulfate (SDS) sample buffer and heating to 100°C for 5 min. Samples were resolved onto 6% or 15% SDS-polyacrylamide gels, stained with Commassie R-250 staining solution (0.5% Coomassie R-250, 25% isopropanol, 10% acetic acid), destained with 50% methanol/5% glycerol, dried and exposed to a PhosphorImager plate (Molecular Dynamics, Piscataway, NJ), and visualized using ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, CA). For the analysis of the phosphorylation of peptides, peptides were applied to individual 2.5 cm-diameter disks of P-81 phosphocellulose filter paper (Schleicher & Schuell) that were immediately immersed in 75 mM phosphoric acid. After extensive wash with 75 mM phosphoric acid, P-81 filters were rinsed with acetone and allowed to air dry. Filters were immersed in Cytoscint liquid scintillation cocktail (Fisher Scientific) and 32P radioactivity on each filter was measured by liquid scintillation using an LS6500 counter (Beckman Coulter).
Results and Discussion
Generation and characterization of active full-length GST-LRRK2
In order to readily generate substantial amounts of purified LRRK2, the full-length enzyme was expressed in 293T cells as an N-terminal tagged GST protein that was purified using glutathione-sepharose beads as described in “Materials and Methods” (Fig. 1A). Since the G2019S mutation is the most common alteration causal of PD and several studies have demonstrated increased kinase activity due to this mutation, the enzyme with this mutation was also produced. To verify the specificity of the purified recombinant GST-LRRK2, the “TDK” kinase dead version of this protein [28] also was assayed. The in-vitro kinase activity of recombinant WT and G2019S GST-LRRK2 was investigated by auto-phosphorylation and phosphorylation of the generic kinase substrate MBP (Fig1A). The G2019S mutation resulted in a 3-5 fold increase in levels of autophosphorylation as well as MBP phosphorylation. The TKD protein demonstrated a paucity of kinase activity. To further confirm the specificity of this activity, it was assessed with the synthetic peptide LRRKtide that contains residue T558 in moesin which was previously identified as a good substrate for LRRK2 [26]. Recombinant WT GST-LRRK2 readily phosphorylated LRRKtide, while the dead mutant showed no activity. In contrast, the activity for the G2019S mutant was ∼10 times higher than for WT in this assay (Fig 1B), suggesting that the effects of the G2019S mutation may be more pronounced than previously believed. The reason(s) for the greater increase in kinase activity by the G2019S mutation using LRRKtide in this system are currently under investigation.
Figure 1. Characterization of recombinant GST-LRRK2 activity.
(A) Western blot analysis with anti-GST or anti-LRRK2 (1181) antibody showing glutathione affinity chromatography purified WT, G2019S and TDK GST-LRRK2 compared to mock transfection. Autoradiography showing that WT and G2019S GST-LRRK2 undergo autophosphorylation and that they phosphorylate MBP, while the TDK mutant is deficient in activity, similar to mock transfection. (B-D) Substrate peptide analysis showing that WT and G2019S GST-LRRK2 specifically phosphorylate LRRKtide, but not a spectrum of other kinase peptide substrates.
To further characterize the in-vitro kinase activity of LRRK2, and screen for other motifs that this enzyme may recognize, the ability of GST-LRRK2 to phosphorylate characterized synthetic peptides with known kinase recognition motifs was analyzed. Some of the peptides studied were based on findings from the inhibitor studies (see below), as well as the ability of LRRK2 to phosphorylate MBP. GST-LRRK2 was assayed for the ability to phosphorylate PKC substrates (MBP fragment 104-118, [Ser25]-PKC fragment 19-31, MBP fragment 4-14, MARCKS PSD-Derived peptide), ERK1/2 substrate (MBP fragment 94-102), PKA substrate (Kemptide), and casein kinase II specific peptide (Figs 1C and D; data not shown). Compared to reactions using LRRKtide, these peptides demonstrated less than 5% of the level of phosphorylation indicating that they are not substrates for LRRK2 (Fig 1C). Similarly, none of these peptides were substrates for G2019S GST-LRRK2 (Fig 1D).
Recombinant GST-LRRK2 was used to screen a range of defined kinase inhibitors
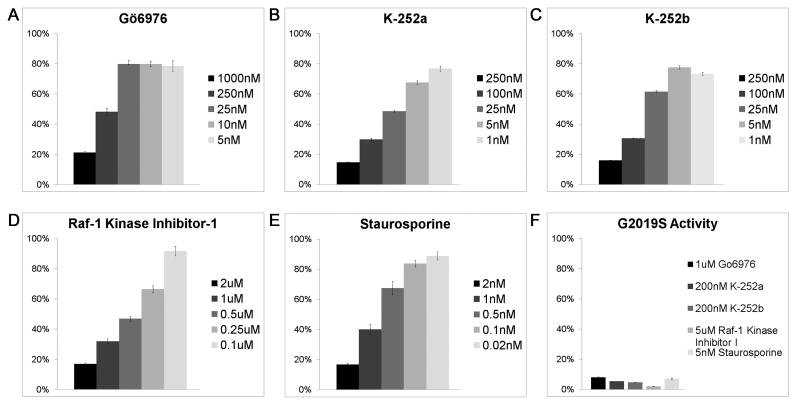
Compounds (listed in “Material and Methods”) specific for different families of kinases including protein kinase A, protein kinase C, mitogen-activated protein kinase, mitogen-activated protein kinase kinase, casein kinase I/II, Ca2+/calmodulin-dependent kinase II, glycogen synthase kinase-3, Rho kinase, Raf kinase, myosin light chain kinase, cyclin-dependent kinases and mixed lineage kinases were assayed using a 1-10 μM range. The majority of compounds had no significant affect on kinase activity (data not shown); however five inhibitors that abolished kinase activity were identified, and were further characterized (Fig 2). Two of these inhibitors inhibited LRRK2 in the high nM range: Raf-1 kinase inhibitor I (IC50 ∼500 nM) (Fig 2D) and Gö6976 (IC50 ∼250nM) (Fig 2A). Three other inhibitors were much more potent, inhibiting LRRK2 in the low nM range: staurosporine (IC50 ∼1nM) (Fig 2E), K252a (IC50 ∼25nM) (Fig 2B), and K252b (IC50 ∼50nM) (Fig 2C). Similarly, all these inhibitors demonstrated the ability to inhibit G2019S LRRK2 (Fig 2F).
Figure 2. Analysis of GST-LRRK2 kinase activity in the presence of various inhibitors.
(A) Gö6976, (B) K-252a, (C) K252-b, (D) Raf-1 kinase inhibitor 1 and (E) Staurosporine are shown to be potent inhibitors of WT GST-LRRK2 using a range of concentration that was used to approximate the IC50. (F) G2019S GST-LRRK2 was also efficiently inhibited by these kinase inhibitors. The values correspond to the phosphorylation of LRRKtide in the presence of each inhibitor relative to reactions without inhibitors expressed as percent activity. (n=3, Error bars indicate standard error)
The four most potent inhibitors of LRRK2 identified here (Gö6976, K252a, K252b and staurosporine) all have a basic indolocarbazole structure: a symmetric fusion of alternating three benzene and two pyrrole rings with a pyrrolidione ring fused to the central benzene structure [31; 32]. These molecules are competitive inhibitors interacting with the ATP binding site of kinases [33-35]. Although staurosporine is a potent kinase inhibitor, it is not very specific [34; 35]. K252a and K252b were identified from the culture broth of the Nocardiopsis bacterium, and although they were initially shown to inhibit PKC [33], they also can inhibit several other kinases [36; 37]. Gö6976 was developed as a specific inhibitor of some PKC isozymes [38], but more recently it was shown to also inhibit some other kinases [39]. The identification of kinases inhibitors that are completely specific has been a challenge, nevertheless once a specific basis structure has been identified several approaches can be used to rationally modify kinase inhibitors to make them more selective ligands [37; 40-42].
The findings described here further support the notion that elevated LRRK2 activity is a cause of pathogenesis in patients with LRRK2 mutations. The identification of potent LRRK2 kinase inhibitors will serve as useful tools for some future in vivo studies of LRRK2 biological function. Collectively, these findings provide important insights and tools for the studies of the mechanisms of LRRK2 pathobiology and could lead to the development of novel therapeutic interventions.
Acknowledgments
This work was funded by grants from the National Institute on Aging (AG09215), the National Institute of Neurological Disorders and Stroke (NS053488) and The Ellison Medical Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Goedert M. Alpha-synuclein and neurodegenerative diseases. Anonymous. 2001:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 2.Forman MS, Lee VM, Trojanowski JQ. Nosology of Parkinson's disease: looking for the way out of a quackmire. Neuron. 2005;47:479–482. doi: 10.1016/j.neuron.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey CP, Giasson BI. Role of mitochondrial dysfunction in Parkinson's disease: Implications for treatment. Drugs Aging. 2007;24:95–105. doi: 10.2165/00002512-200724020-00002. [DOI] [PubMed] [Google Scholar]
- 4.Cookson MR. The biochemistry of Parkinson's disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 5.Dawson TM, Dawson VL. Molecular Pathways of Neurodegeneration in Parkinson's Disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 6.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 7.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 8.Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, Romito L, Albanese A, Dallapiccola B, Bentivoglio AR. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56:336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 9.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 10.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 Cause Autosomal-Dominant Parkinsonism with Pleomorphic Pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der BM, de Munain AL, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the Gene Containing Mutations that Cause PARK8-Linked Parkinson's Disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Giasson BI, Van D V. Mutations in LRRK2 as a cause of Parkinson's disease. Neurosignals. 2008;16:99–105. doi: 10.1159/000109764. [DOI] [PubMed] [Google Scholar]
- 13.Paisan-Ruiz C, Lang AE, Kawarai T, Sato C, Salehi-Rad S, Fisman GK, Al Khairallah T, George-Hyslop P, Singleton A, Rogaeva E. LRRK2 gene in Parkinson disease. Neurology. 2005;65:696–700. doi: 10.1212/01.wnl.0000167552.79769.b3. [DOI] [PubMed] [Google Scholar]
- 14.Deng H, Le W, Guo Y, Hunter CB, Xie W, Jankovic J. Genetic and clinical identification of Parkinson's disease patients with LRRK2 G2019S mutation. Ann Neurol. 2005;57:933–934. doi: 10.1002/ana.20510. [DOI] [PubMed] [Google Scholar]
- 15.Di Fonzo A, Rohe CF, Ferreira J, Chien HF, Vacca L, Stocchi F, Guedes L, Fabrizio E, Manfredi M, Vanacore N, Goldwurm S, Breedveld G, Sampaio C, Meco G, Barbosa E, Oostra BA, Bonifati V. A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson's disease. Lancet. 2005;365:412–415. doi: 10.1016/S0140-6736(05)17829-5. [DOI] [PubMed] [Google Scholar]
- 16.Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, Payami H, Nutt J, Maraganore DM, Czyzewski K, Styczynska M, Wszolek ZK, Farrer MJ, Toft M. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76:672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, Singleton A, Foroud T. Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 18.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 19.Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 20.Lesage S, Ibanez P, Lohmann E, Pollak P, Tison F, Tazir M, Leutenegger AL, Guimaraes J, Bonnet AM, Agid Y, Durr A, Brice A. G2019S LRRK2 mutation in French and North African families with Parkinson's disease. Ann Neurol. 2005;58:784–787. doi: 10.1002/ana.20636. [DOI] [PubMed] [Google Scholar]
- 21.Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, Lipton RB, Soto-Valencia J, Risch N, Bressman SB. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 22.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 Cause Autosomal-Dominant Parkinsonism with Pleomorphic Pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der BM, de Munain AL, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the Gene Containing Mutations that Cause PARK8-Linked Parkinson's Disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 24.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 26.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 30.Chen CA, Okayama H. High-efficient transfection using calcium phosphate-DNA precipitate formed in BES. In: Ausubel FEA, editor. Currents Protocols in Molecular Biology. John Wiley & Sons, Inc.; New York: 1997. pp. 917–919. [Google Scholar]
- 31.Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 32.Ross AH, McKinnon CA, Daou MC, Ratliff K, Wolf DE. Differential biological effects of K252 kinase inhibitors are related to membrane solubility but not to permeability. J Neurochem. 1995;65:2748–2756. doi: 10.1046/j.1471-4159.1995.65062748.x. [DOI] [PubMed] [Google Scholar]
- 33.Kase H, Iwahashi K, Nakanishi S, Matsuda Y, Yamada K, Takahashi M, Murakata C, Sato A, Kaneko M. K-252 compounds novel and potent inhibitors of protein kinase C and cyclic nucleotide-dependent protein kinases. Biochem Biophys Res Commun. 1987;142:436–440. doi: 10.1016/0006-291x(87)90293-2. [DOI] [PubMed] [Google Scholar]
- 34.Fabian MA, Biggs WH, III, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 35.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 36.Hidaka H, Kobayashi R. Pharmacology of protein kinase inhibitors. Annu Rev Pharmacol Toxicol. 1992;32:377–397. doi: 10.1146/annurev.pa.32.040192.002113. [DOI] [PubMed] [Google Scholar]
- 37.Wang LH, Besirli CG, Johnson EM., Jr Mixed-lineage kinases: a target for the prevention of neurodegeneration. Annu Rev Pharmacol Toxicol. 2004;44:451–474. doi: 10.1146/annurev.pharmtox.44.101802.121840. [DOI] [PubMed] [Google Scholar]
- 38.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 39.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crespo A, Zhang X, Fernandez A. Redesigning kinase inhibitors to enhance specificity. J Med Chem. 2008;51:4890–4898. doi: 10.1021/jm800453a. [DOI] [PubMed] [Google Scholar]
- 41.Le CS, Klafki HW, Plesnila N, Hubinger G, Obermeier A, Sahagun H, Monse B, Seneci P, Lewis J, Eriksen J, Zehr C, Yue M, McGowan E, Dickson DW, Hutton M, Roder HM. An inhibitor of tau hyperphosphorylation prevents severe motor impairments in tau transgenic mice. Proc Natl Acad Sci U S A. 2006;103:9673–9678. doi: 10.1073/pnas.0602913103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bregman H, Carroll PJ, Meggers E. Rapid access to unexplored chemical space by ligand scanning around a ruthenium center: discovery of potent and selective protein kinase inhibitors. J Am Chem Soc. 2006;128:877–884. doi: 10.1021/ja055523r. [DOI] [PubMed] [Google Scholar]