Abstract
Familial dysautonomia (FD) is a hereditary neuronal disease characterized by poor development and progressive degeneration of the sensory and autonomic nervous system. Majority of FD (99.5%) results from a single nucleotide point mutation in the IKBKAP gene encoding IKAP, also known as elongation protein 1 (ELP1). The point mutation leads to variable, tissue specific expression of a truncated IKBKAP mRNA. The appearance of the truncated IKBKAP coincides with a marked reduction of its wild type mRNA leading to decreased IKAP protein levels especially in the sensory and autonomous nervous system. Recently, two independent studies were carried out to establish a cellular model system to study the loss-of-function of IKAP in mammalian cells. Both studies used RNA interference to deplete wild type IKAP from different mammalian cell types. In both studies the depletion of IKAP resulted in a cell migration defect, revealing the importance of IKAP in this process. These studies lead to a common conclusion according to which defective neuronal migration could underlie FD. They gave however two very different explanations of how IKAP would regulate cell migration: via transcriptional regulation and via cytosolic interactions.
Key words: actin, elongation protein 1, familial dysautonomia, filamin A, IKAP, c-Jun N-terminal kinase, leading edge, membrane ruffle, migration
IKAP is a well-conserved 150 kDa eukaryotic protein that does not belong to any protein family. It lacks a clear domain structure that could be used to predict its biological function. Even though the vast majority (over 90%) of IKAP resides in the cytosol,1–4 it is best known and characterized as a member of the human holo-Elongator complex involved in RNA polymerase II-mediated transcription elongation in the nucleus. This complex consists of three proteins, IKAP/ELP1, ELP2 and ELP3, of which ELP3 is an acetyltransferase that acetylates histones H3 and H4.5 In addition to nucleus, IKAP can be co-precipitated with ELP2 and ELP3 also from cytosol suggesting that the complex could have a functional role there as well.4 Several cytosolic functions have been thus far suggested for IKAP itself such as the regulation of c-Jun N-terminal kinase (JNK) signaling,1 exocytosis3 and tRNA modification,6 clearly pointing out for the multi functionality of IKAP as well as further challenging the efforts to understand the molecular basis of FD.
FD is an autosomal recessive congenital neuropathy with a carrier frequency of 1 in 30 for Ashkenazi Jews. FD is characterized by poor development, poor maintenance and progressive degeneration of the sensory and autonomous nervous system.7 Vast majority of patients suffering from FD exhibit a single point mutation in IKBKAP gene encoding IKAP, leading to a variable exon skipping and tissue specific expression of a truncated IKBKAP transcript.8,9 It is not really known how this abnormal splicing of IKBKAP results in deficient development and maintenance and progressive degeneration of sensory and autonomous nervous system. According to the existing theory the resulting loss of the wild type IKAP, that is most extensive and thus especially destructive for the sensory and autonomous nerves, would be responsible of FD.
We recently utilized the RNA interference technology to study how various mammalian cells, including primary neurons, would respond to the loss of IKAP.4 Similar study was conducted earlier, however only concentrating on the transcriptional differences between IKAP depleted and corresponding control cells.10 These studies reported significant migration4,10 and adhesion4 defect in cells expressing very low amounts (approximately 20%) of IKAP, thus identifying as well as verifying functional importance of IKAP in cell migration (Fig. 1). We showed that these defects were clearly associated with inability of filamin A to localize to the leading edges of the migrating cells as well as with disorganized actin cytoskeleton (Fig. 1).4 We also demonstrated that all these defects could be fully rescued by co-expression of wild type IKAP unrecognizable for RNA interference tools but not by FD-IKAP, a truncated IKAP made according to the major mutation found in FD, clearly suggesting that the wild type IKAP would be needed for these processes. Furthermore, we found that the cytosolic pool of IKAP associated with filamin A as well as with several proteins involved in cell motility such as dynein heavy chain, the ubiquitin-specific processing protease USP9X, the non-receptor tyrosine kinase BMX, ELP2 and ELP3, which further supports the role of the cytosolic associations of IKAP in cell adhesion and migration.4
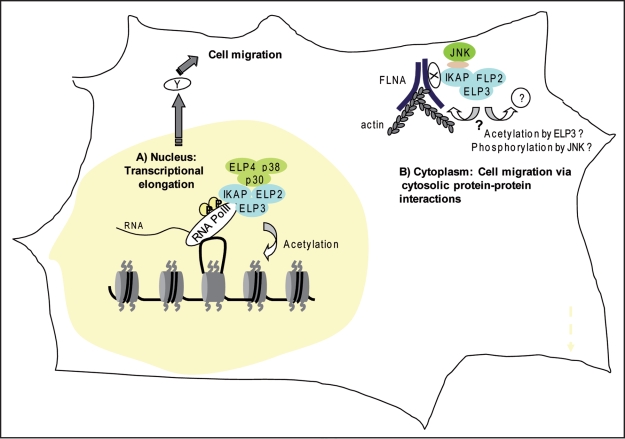
Figure 1.
The two described cellular functions of IKAP in mammalian cells. (A) In the nucleus IKAP is part of the core-elongator complex (IKAP, ELP2, ELP3, ELP4, p30 and p38) that binds hyperphosphorylated RNA polymerase II and assists it in transcriptional elongation by acetylating histones. There is contradictory evidence on whether IKAP/elongator regulates the expression of paxillin, beclin1, alpha-tubulin or other genes whose protein products (collectively called here as protein Y) regulate cell migration in the cytoplasm. (B) In the cytoplasm IKAP associates among others with ELP2, ELP3 and c-Jun N-terminal kinase (JNK) and indirectly (through protein X) with filamin A (FLNA) and has a role in cell migration. IKAP regulates the localization of filamin A to membrane ruffles and actin cytoskeleton organization, possibly through filamin A. It is not clear whether ELP4, p38 and p30 are also part of the complex in the cytoplasm and whether the complex has acetylase activity. It is possible that the complex regulates actin cytoskeleton and cell migration by acetylating and/or phosphorylating filamin A or other cytoplasmic targets.
Consistent with the role of IKAP in transcriptional elongation, the earlier study showed that the migration defect that results from the depletion of IKAP would be due to defect in transcriptional elongation. In the study this defect was found to be targeted to a set of genes involved in cell migration, of which paxillin, beclin 1 and alpha tubulin were the most affected.10 There are several reasons however, that substantially question this conclusion. First of all the results were based on a single RNA interferences sequence.11 Sequence analysis with the Dharmacon off-target engine (www.dharmacon.com/seedlocator/default.aspx) of this interfering sequence identifies both paxillin (multiple seed matches) and beclin 1 (at least one seed match) as its possible off-targets. Secondly, in our study which was based on several different IKAP RNA interference sequences, we were unable to detect changes in the protein levels of paxillin, beclin 1 or tubulin despite of a fact that we also identified severe adhesion and migration deficiency in the cells. Thirdly, we also did expression array analysis utilizing a different IKAP RNA interfering sequence to investigate the expression differences of a selected set of genes involved in cell adhesion and migration, and found no significant differences in their mRNA levels in comparison to control cells (except for NCAM, which difference was not however translated to its protein level).4 Fourthly, expression array analysis of cerebrum samples of FD patients failed to show changes in paxillin, beclin 1, tubulin or other mRNAs related to adhesion or migration in comparison to control individuals.12 All this together casts some doubt for transcription elongation problems, at least these particular genes or others commonly related to adhesion and migration, as the main cause of FD or as the cause of the adhesion and migration deficiencies detected in IKAP depleted cells (Fig. 1). Considering the importance of transcriptional elongation for various cellular processes and the efficient association of IKAP with ELP2 and ELP3, it is reasonable to assume that cells would have a mechanism that ensures the small nuclear pool of the holo-Elongator in any conditions. In this respect techniques such as RNA interference, which in case of IKAP leads at maximal of 80% depletion, might not suffice to identify IKAP target genes.
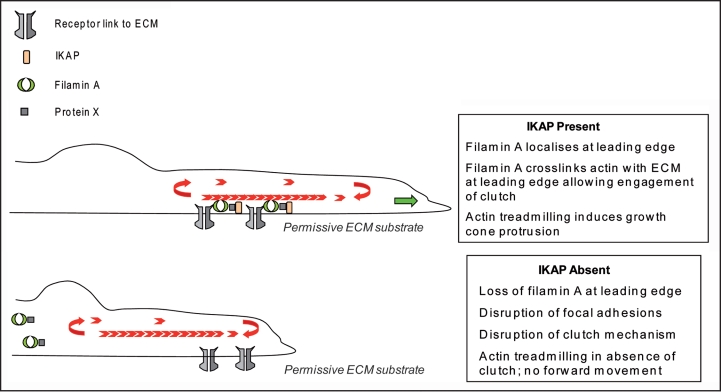
How can the cytosolic interactions of IKAP regulate cell adhesion and migration and perhaps even contribute to FD? Even though we and others have characterized several possible players in the IKAP-mediated cell adhesion and migration, the exact mechanism still awaits to be discovered. Thus we can currently only speculate about it. Our current model is based on the association of IKAP and filamin A, which seems as the most promising IKAP-mediated cytosolic interaction in respect to cell migation (Fig. 2). For example, the fact that IKAP associates with and is needed for the membrane ruffle localization of filamin A, a protein capable of driving cell migration by reorganizing the actin cytoskeleton and generating forces necessary for cell motility,13 suggests that the interplay between IKAP and filamin A is important for proper migration of cells. Interestingly, loss-of-function mutations in human filamin A cause a neurodevelopmental disease, periventricular heterotropia (PH). The disease is believed to be result from a defect in the migration of postmitotic neurons due to inability to properly crosslink and mobilize actin, as the mutations observed in filamin A often involve truncation or disruption of its actinbinding domain.13 In support of this, in mice the expression of filamin A lacking the actin-binding domain inhibits the migration of postmitotic neurons14 and cells lacking filamin A are unable to migrate until rescued by ectopic expression of filamin A.15 Since depletion of IKAP also lead in disorganized actin cytoskeleton, and actin-filamin A interactions are important for neuronal migration, we propose a model of how IKAP could regulate neuronal migration via filamin A association. This model is based on the “slippage clutch mechanism”-model,16,17 whereby localization of filamin A at the leading edge would engage the “clutch mechanism” and allow actin treadmilling resulting in forward movement of the growth cone. In the absence of IKAP, filamin A is mislocalized and therefore the clutch is not engaged (Fig. 2).
Figure 2.
Theoretical model for the function of IKAP in neuronal migration. IKAP is required for neurons to migrate. We have shown that filamin A localiszation to the leading edges of migrating cells requires the presence of IKAP. We suggest that IKAP plays a similar function in neurons, where it would recruit filamin A to the leading edge of the growth cone via an undefined interaction partner (protein X). Here filamin A functions to crosslink the extracellular matrix (ECM) to the actin cytoskeleton. Thus appropriate localisation of filamin A at the leading edge engages a “clutch” mechanism. When the clutch is engaged, actin treadmilling generates a protrusive force at the leading edge resulting in forward movement of the growth cone. In the absence of IKAP, filamin A is mislocalized and therefore the clutch is not engaged. In this context, there is futile actin treadmilling, no protrusive force is generated and the neuron does not move forward.
How could IKAP regulate filamin A? In our model (Fig. 2) IKAP would function in the cytosol by physically guiding filamin A close to actin and into the leading edges of migrating cells. Theoretically IKAP could also induce post-translational modifications that would promote its localization to membrane ruffles. In either case, it seems that IKAP is not directly binding to filamin A since their interaction could not be seen in vitro. In case of possible post-translational modifications, IKAP does not contain any catalytic activity to directly modify other proteins. However, IKAP could induce the modification of filamin A or its associating protein(s) by bringing it in close proximity with other proteins possessing catalytic activity such as ELP3 or JNK that can induce acetylation or phosphorylation, respectively. Indeed, cytoskeletal proteins and regulators of cytoskeleton organization, like tubulin, actin and cortactin, are regulated by acetylation. Moreover, histone deacetylase (HDAC) inhibitor treatment results in accumulation of stabilized actin stress fibers whereas HDAC agonist treatment potently inhibits stress fiber formation and/or stability in tissue culture conditions.19 On the other hand it has been shown in yeast that the whole core-elongator is needed for its histone acetylase activity,20 and thus it is not known if ELP3 could acetylate proteins outside the core-elongator. Interestingly, the carboxy-terminus of IKAP however seems to be especially important fort its migratory properties since the expression of the amino-terminal fragment of IKAP (FD-IKAP) could not rescue IKAP depleted cells but the expression of the carboxy-terminal fragment, which is missing from the defectively spliced IKAP present in FD, gave significant although partial rescue (Johansen LD and Kallunki T, unpublished observations).
It would be very important to find out if ELP3 localizes to membrane ruffles as well and whether its depletion leads to migration defects. Respectively, it would also be relevant to investigate whether filamin A or its associating proteins can be acetylated by ELP3. In respect to JNK, it was recently reported that an upstream kinase of JNK-pathway, MEKK4 (MAP3K4) is involved in the regulation of filamin A by both modulating filamin A expression and also by associating with filamin A, and that inhibition of MEKK4 activity leads to inhibition of neuronal migration.18 JNK has also been reported to phosphorylate microtubule associated protein doublecortin (DCX) in growth cones of migrating neurons.21 DCX can interact with the ubiquitin specific protease (USP9X)22 and dynein heavy chain,23 both of which also associated with IKAP in our study, suggesting that local changes in the activation of JNK pathway could also be involved in IKAP-mediated migration.
The outgrowth of axons during the development, as well as in the regeneration of the nervous system after injury, is controlled by extracellular cues. Most extracellular cues initiate signaling cascades that converge onto cytoskeleton and thus cytoskeleton and cytoskeleton-associated proteins can modulate the neuronal guidance process.24,25 The defects in these molecules can affect both the development as well as maintenance of the nervous system as in the latter case has been shown for PH. In this respect the defect cytoskeletal organization and filamin A localization resulting from the loss-of-function of IKAP could indeed underlie or at least contribute to the pathology of FD. It is good to remember that even though our current work concentrated on filamin A, which we found most interesting of the identified IKAP association partners in respect to neuronal migration, we did identify many others that might turn even more important in respect to FD. Furthermore, the dominant inhibitory effect of the FD-IKAP to neuronal migration we reported in the article could also partially contribute to FD.4 Even though huge progress towards the understanding of the function of IKAP in respect to FD have been made during the years passed since the identification of IKBKAP mutations as the cause of FD in 2001, the detailed mechanism of the function of IKAP and its role in the development of FD still awaits to be clarified.
Acknowledgements
Danish Cancer Society, Dysautonomia Foundation Inc., Danish Medical Research Council and Danish National Research Foundation.
Abbreviations
- BMX
bone marrow X kinase
- DCX
doublecortin
- ELP1
elongation protein 1
- ELP2
elongation protein 2
- ELP3
elongation protein 3
- FD
familial dysautonomia
- HDAC
histone deacetylase
- IKAP
IkappaB kinase associated protein
- JNK
c-Jun N-terminal kinase
- PH
periventricular heterotropia
Note
When analyzing the IKAP siRNA oligo (ref. 11) in the Dharmacon off target engine (www.dharmacon.com/seedlocator/default.aspx) one needs to omit the two first AA:s (the leader sequence) from the oligo and use the following 19 nucleotides only. It is however widely accepted to begin the siRNA oligo with the AA-leader sequence when reporting the sequence.
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/6630
References
- 1.Holmberg C, Katz S, Lerdrup M, Herdegen T, Jäättelä M, Aronheim A, et al. A novel specific role for IkappaB kinase complex-associated protein in cytosolic stress signaling. J Biol Chem. 2002;277:31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 2.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 3.Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Johansen LD, Naumanen T, Knudsen A, Westerlund N, Gromova I, Junttila M, et al. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J Cell Sci. 2008;121:854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 5.Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 6.Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Axelrod FB. Familial dysautonomia. Muscle Nerve. 2004;29:352–363. doi: 10.1002/mus.10499. [DOI] [PubMed] [Google Scholar]
- 8.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 11. [IKAP target sequence for the siRNA used in the Close et al., (ref. 10): AATCCTCAGTGCTTCTCTCTC; available from authors on request and discussed in Johansen et al., (ref. 4).]
- 12.Cheishvili D, Maayan C, Smith Y, Ast G, Razin A. IKAP/hELP1 deficiency in the cerebrum of familial dysautonomia patients results in down regulation of genes involved in oligodendrocyte differentiation and in myelination. Hum Mol Genet. 2007;16:2097–2104. doi: 10.1093/hmg/ddm157. [DOI] [PubMed] [Google Scholar]
- 13.Sarkisian MR, Bartley CM, Rakic P. Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex. Trends Neurosci. 2008;31:54–61. doi: 10.1016/j.tins.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Nagano T, Morikubo S, Sato M. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J Neurosci. 2004;24:9648–9657. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–357. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 16.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang YL. Flux at focal adhesions: slippage clutch, mechanical gauge or signal depot. Sci STKE. 2007;377:10. doi: 10.1126/stke.3772007pe10. [DOI] [PubMed] [Google Scholar]
- 18.Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, Torii M, et al. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA. 2002;9:2352–3517. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, Fishler Y, Azoulai D, Reiner O. DCX, a new mediator of the JNK pathway. EMBO J. 2004;23:823–832. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friocourt G, Kappeler C, Saillour Y, Fauchereau F, Rodriguez MS, Bahi N, et al. Doublecortin interacts with the ubiquitin protease DFFRX, which associates with microtubules in neuronal processes. Mol Cell Neurosci. 2005;28:153–164. doi: 10.1016/j.mcn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent EW, Tang F, Kalil K. Axon guidance by growth cones and branches: common cytoskeletal and signaling mechanisms. Neuroscientist. 2003;9:343–353. doi: 10.1177/1073858403252683. [DOI] [PubMed] [Google Scholar]
- 25.Bouquet C, Nothias F. Molecular mechanisms of axonal growth. Adv Exp Med Biol. 2007;621:1–16. doi: 10.1007/978-0-387-76715-4_1. [DOI] [PubMed] [Google Scholar]