Abstract
Junctional Adhesion Molecule A (JAM-A) is a member of the Ig superfamily of membrane proteins expressed in platelets, leukocytes, endothelial cells and epithelial cells. We have previously shown that in endothelial cells, JAM-A regulates basic fibroblast growth factor, (FGF-2)-induced angiogenesis via augmenting endothelial cell migration. Recently, we have revealed that in breast cancer cells, downregulation of JAM-A enhances cancer cell migration and invasion. Further, ectopic expression of JAM-A in highly metastatic MDA-MB-231 cells attenuates cell migration, and downregulation of JAM-A in low-metastatic T47D cells enhance migration. Interestingly, JAM-A expression is greatly diminished as breast cancer disease progresses. The molecular mechanism of this function of JAM-A is beyond its well-characterized barrier function at the tight junction. Our results point out that JAM-A differentially regulates migration of endothelial and cancer cells.
Key words: JAM-A, integrin, αvβ3, FGF-2, breast cancer, cell migration and invasion, T47D, MDA-MB-231, siRNA
Endothelial and epithelial cells exhibit cell polarity and have characteristic tight junctions (TJs) that separate apical and basal surfaces. TJs are composed of both transmembrane and cytoplasmic proteins. The three major families of transmembrane proteins include claudins, occludin and JAM family members.1–3 Additionally, interaction between the peripheral proteins such as PDS-95/Discs large/ZO family (PDZ) domain-containing proteins in TJs plays an important role in maintaining the junctional integrity.2,4,5
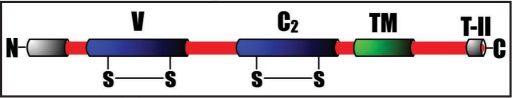
JAMs are type I membrane proteins (Fig. 1) predominately expressed in endothelial and epithelial cell TJs, platelets and some leukocytes.6–8 The classical JAMs are JAM-A, JAM-B and JAM-C, which can all regulate leukocyte-endothelial cell interaction through their ability to undergo heterophilic binding with integrins αLβ2 or αvβ3, α4β1 and αMβ2 respectively. The cytoplasmic tail of JAMs contains a type II PDZ-domain-binding motif (Fig. 1) that can interact with the PDZ domain containing cytoplasmic molecules such as ZO-1, ASIP/PAR-3 or AF-6.9,10 Additionally, consistant with their junctional localization and their tendency to be involved in homophilic interactions, JAMs have been shown to modulate paracellular permeability and thus may play an important role in regulating the epithelial and endothelial barrier.11,12 In addition, ectopic expression of JAM-A in CHO cells promotes localization of ZO-1 and occludin at points of cell contacts, which suggests a role for JAM-A in TJ assembly.10,13,14 Recently, it has been shown that JAM-A regulates epithelial cell morphology by modulating the activity of small GTPase Rap1 suggesting a role for JAM-A in intracellular signaling.15
Figure 1.
Schematic representation of the domain structure of JAM family proteins. V, variable Ig domain; C2, constant type 2 Ig domain; TM, transmembrane domain; T-II, Type II PDZ-domain binding motif.
We have previously shown that JAM-A is a positive regulator of fibroblast growth factor-2 (FGF-2) induced angiogenesis.16 Evidence was provided to support the notion that JAM-A forms a complex with integrin αvβ3 at the cell-cell junction in quiescent human umbilical cord vein endothelial cells (HUVECs) and FGF-2 dissociates this complex.16 It was further established that inhibition of JAM-A using a function-blocking antibody also inhibits FGF-2 induced HUVECs migration in vitro and angiogenesis in vivo. Overexpression of JAM-A induced a change in HUVECs morphology similar to that observed when treated with FGF-2.17 Furthermore, overexpression of JAM-A, but not its cytoplasmic domain deletion mutant, augmented cell migration in the absence of FGF-2.17 In addition, downregulation of JAM-A in HUVECs using specific siRNA, resulted in reduced FGF-2-induced cell migration and inhibition of mitogen activated protein (MAP) kinase activation.18 These findings clearly suggested that JAM-A positively regulates FGF-2-induced endothelial cell migration. This was further confirmed in vivo by using JAM-A null mouse in which FGF-2 failed to support angiogenesis.19
It is known that JAM-C, a JAM family member, is involved in the process of tumor cell metastasis.20 However, little is known about JAM-A's role in cancer progression. We recently found that JAM-A is expressed in breast cancer tissues and cell lines.21 Based on our studies with endothelial cells it was felt that JAM-A expression in breast cancer cells may also enhance the migratory ability of these cells. Surprisingly, we found an inverse relation between the expression of JAM-A and the metastatic ability of breast cancer cells. T47D cells, which express high levels of JAM-A, are the least migratory; whereas MDA-MB-231 cells, which are highly migratory, are found to express the least amount of JAM-A.21 We also found that overexpression of JAM-A in MDA-MB-231 cells caused a change in cell morphology from spindle-like to rounded shape and formed cobblestone-like clusters.21 This is consistent with the previous report, that downregulation of JAM-A expression from epithelial cells using siRNA results in the change of epithelial cell morphology.15 This change in cell morphology by knockdown of JAM-A was attributed to the disruption of epithelial cell barrier function.15 It was further shown that knockdown of JAM-A affects epithelial cell morphology through reduction of β1integrin expression due to decreased Rap1 activity.15 Our observed effect of JAM-A downregulation in T47D cells, however, is not due to downregulation of β1integrin, since the level of this integrin was not affected in these cells. Interestingly, overexpression of JAM-A significantly affected both the cell migration and invasion of MDA-MB-231 cells. Furthermore, knockdown of JAM-A using siRNA enhanced invasiveness of MDA-MB-231 cells, as well as T47D cells.21 The ability of JAM-A to attenuate cell invasion was found to be due to the formation of functional tight junctions as observed by distinct accumulation of JAM-A and ZO-1 at the TJs and increased transepithelial resistance. These results identify, for the first time, a tight junctional cell adhesion protein as a key negative regulator of breast cancer cell migration and invasion.21
JAM-A has been shown to be important in maintaining TJ integrity.15,22–25 Disruption of TJs has been implicated to play a role in cancer cell metastasis by inducing epithelial mesenchymal transition.26 Several laboratories, including ours, have shown that cytokines and growth factors redistribute JAM-A from TJs.16,27,28 Consistent with this finding, it has been shown that hepatocyte growth factor (HGF) disrupts TJs in human breast cancer cells and downregulates expression of several TJ proteins.29 It is therefore conceivable that the loss of JAM-A in highly metastatic cells is a consequence of disruption of TJs. This was further supported by the findings that overexpression of JAM-A forms functional TJs in MDA-MB-231 cells and attenuates their migratory behavior. Our result is the first report correlating an inverse relationship of JAM-A expression in breast cancer cells to their invasive ability.21
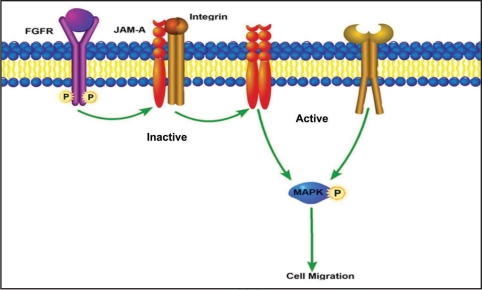
Using cDNA microarray technology, it has been revealed how genes involved in cell-cell adhesion, including those of the TJ, are under or overexpressed in different carcinomas.15,30 Cell-cell adhesion molecules have been well documented to regulate cancer cell motility and invasion. Of these, the cadherin family have been studied the most.31,32 It was proposed that a cadherin switch, that is, the loss of E-cadherin and subsequent expression of N-cadherin, may be responsible for breast cancer cell invasion.33,34 Although the role of cadherins is well-documented, it remains controversial since some breast cancer cell lines that do not express these proteins still posses highly invasive characteristics.33,34 However, the observed effect of overexpression of JAM-A does not appear to be simply due to the formation of TJs, since individual cells that express increased JAM-A show reduced migration.21 This is not surprising, considering the fact that JAM-A in addition to its function of regulating TJ integrity is also shown to participate in intracellular signaling. JAM-A is capable of interacting homotypically as well as heterotypically on the cell surface.35,36 It has also been shown that it interacts with several cytoplasmic proteins through its PDZ domain-binding motif and recruits signaling proteins at the TJs.37 Recent findings using site-directed mutagenesis suggest that cis-dimerization of JAM-A is necessary for it to carry out its biological functions.38 Our own observations suggest that a JAM-A function-blocking antibody inhibits focal adhesion formation in endothelial cells (unpublished data), whereas overexpresion of JAM-A in MDA-MB-231 cells show increased and stable focal adhesions.21 It is therefore conceivable that in quiescent endothelial/epithelial cells JAM-A associates with integrin to form an inactive complex at the TJ (Fig. 2). Growth factors such as FGF-2 signaling dissociates this complex thus allowing dimerization of JAM-A and activation of integrin augmenting cell migration (Fig. 2). On the contrary, in MDA-MB-231 cancer cells, which express low levels of JAM-A and do not form tight junctions, there may not be efficient inactive complex formation between JAM-A and integrin. Overexpression of JAM-A in these cells however, may promote such inactive complex formation leading to inhibition of integrin activation and JAM-A dimerization, both necessary events for cell migration. We are currently in the process of determining the specificity of interaction of JAM-A with integrins. Further experimentation is ongoing to determine the contribution of JAM-A dependent signaling in cell migration.
Figure 2.
Schematic representation of JAM-A regulation of cell migration. JAM-A forms an inactive complex with the integrin and sequesters it at the TJs. Growth factor signaling dissociates this complex, promoting integrin activation and JAM-A dimerization leading to cell migration via MAP kinase activation. Ectopic expression of JAM-A in cancer cells may induce its association with integrin, forming an inactive complex and hence attenuation of migration.
JAM-A differentially regulates cell migration in endothelial and cancer cells due to its ability to form inactive complex with integrin, making it a metastasis suppressor. The downregulation of JAM-A in carcinoma cells may be detrimental to the survival of breast cancer patients. It is therefore very important to determine the molecular determinants that are responsible for the downregulation of JAM-A during cancer progression. Thus, JAM-A, a molecule that dictates breast cancer cell invasion, could be used as a prognostic marker for metastatic breast cancer.
Acknowledgements
This work is supported by grants (U.P.N.) from the National Institutes of Health (HL63960) and the National Center for Research Resources (1P20RR155801).
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/6753
References
- 1.Fanning AS, Anderson JM. Protein modules as organizers of membrane structure. Curr Opin Cell Biol. 1999;11:432–439. doi: 10.1016/S0955-0674(99)80062-3. [DOI] [PubMed] [Google Scholar]
- 2.Faris M, Ensoli B, Kokot N, Nel AE. Inflammatory cytokines induce the expression of basic fibroblast growth factor (bFGF) isoforms required for the growth of Kaposi's sarcoma and endothelial cells through the activation of AP-1 response elements in the bFGF promoter. Aids. 1998;12:19–27. doi: 10.1097/00002030-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 4.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik UP, Eckfeld K. Junctional adhesion molecule 1 (JAM-1) J Biol Regul Homeost Agents. 2003;17:341–347. [PubMed] [Google Scholar]
- 9.Ebnet K, Schulz CU, Meyer ZuBrickwedde MK, Pendl GG, Vestweber D. Junctional Adhesion Molecule (JAM) interacts with the PDZ domain containing proteins AF-6 and ZO-1. J Biol Chem. 2000;275:27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- 10.Bazzoni G, Martinez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E. Interaction of Junctional Adhesion Molecule with the Tight Junction Components ZO-1, Cingulin and Occludin. J BiolChem. 2000;275:20520–20526. doi: 10.1074/jbc.M905251199. [DOI] [PubMed] [Google Scholar]
- 11.Kostrewa D, Brockhaus M, D'Arcy A, Dale GE, Nelboeck P, Schmid G, et al. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. Embo J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams LA, Martin-Padura I, Dejana E, Hogg N, Simmons DL. Identification and characterisation of human Junctional Adhesion Molecule (JAM) Mol Immunol. 1999;36:1175–1188. doi: 10.1016/s0161-5890(99)00122-4. [DOI] [PubMed] [Google Scholar]
- 13.Naik UP, Naik MU, Eckfeld K, Martin-DeLeon P, Spychala J. Characterization and chromosomal localization of JAM-1, a platelet receptor for a stimulatory monoclonal antibody. J Cell Sci. 2001;114:539–547. doi: 10.1242/jcs.114.3.539. [DOI] [PubMed] [Google Scholar]
- 14.Soler AP, Miller RD, Laughlin KV, Carp NZ, Klurfeld DM, Mullin JM. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20:1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 15.Mandell KJ, Babbin BA, Nusrat A, Parkos CA. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta1 integrins and Rap1 activity. J BiolChem. 2005;280:11665–11674. doi: 10.1074/jbc.M412650200. [DOI] [PubMed] [Google Scholar]
- 16.Naik MU, Mousa SA, Parkos CA, Naik UP. Signaling through JAM-1 and alphavbeta3 is required for the angiogenic action of bFGF: dissociation of the JAM-1 and alphavbeta3 complex. Blood. 2003;102:2108–2114. doi: 10.1182/blood-2003-04-1114. [DOI] [PubMed] [Google Scholar]
- 17.Naik MU, Naik UP. Junctional adhesion molecule-A-induced endothelial cell migration on vitronectin is integrin {alpha}v{beta}3 specific. J Cell Sci. 2006;119:490–499. doi: 10.1242/jcs.02771. [DOI] [PubMed] [Google Scholar]
- 18.Naik MU, Vuppalanchi D, Naik UP. Essential role of junctional adhesion molecule-1 in basic fibroblast growth factor-induced endothelial cell migration. Arterioscler Thromb Vasc Biol. 2003;23:2165–2171. doi: 10.1161/01.ATV.0000093982.84451.87. [DOI] [PubMed] [Google Scholar]
- 19.Cooke VG, Naik MU, Naik UP. Fibroblast growth factor-2 failed to induce angiogenesis in junctional adhesion molecule-A-deficient mice. Arterioscler Thromb Vasc Biol. 2006;26:2005–2011. doi: 10.1161/01.ATV.0000234923.79173.99. [DOI] [PubMed] [Google Scholar]
- 20.Santoso S, Orlova VV, Song K, Sachs UJ, Andrei-Selmer CL, Chavakis T. The homophilic binding of junctional adhesion molecule-C mediates tumor cell-endothelial cell interactions. J Biol Chem. 2005;280:36326–36333. doi: 10.1074/jbc.M505059200. [DOI] [PubMed] [Google Scholar]
- 21.Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res. 2008;68:2194–2203. doi: 10.1158/0008-5472.CAN-07-3057. [DOI] [PubMed] [Google Scholar]
- 22.Balda MS, Matter K. Tight junctions. J Cell Sci. 1998;111:541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- 23.Balda MS, Matter K. Transmembrane proteins of tight junctions. Semin Cell Dev Biol. 2000;11:281–289. doi: 10.1006/scdb.2000.0177. [DOI] [PubMed] [Google Scholar]
- 24.Bazzoni G. Endothelial tight junctions: permeable barriers of the vessel wall. ThrombHaemost. 2006;95:36–42. [PubMed] [Google Scholar]
- 25.Dejana E, Corada M, Lampugnani MG. Endothelial cell-to-cell junctions. Faseb J. 1995;9:910–918. [PubMed] [Google Scholar]
- 26.Gumbiner BM. Regulation of cadherin adhesive activity. J Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozaki H, Ishii K, Horiuchi H, Arai H, Kawamoto T, Okawa K, et al. Cutting edge: combined treatment of TNFalpha and IFNgamma causes redistribution of junctional adhesion molecule in human endothelial cells. J Immunol. 1999;163:553–557. [PubMed] [Google Scholar]
- 28.Shaw SK, Perkins BN, Lim YC, Liu Y, Nusrat A, Schnell FJ, et al. Reduced expression of junctional adhesion molecule and platelet/endothelial cell adhesion molecule-1 (CD31) at human vascular endothelial junctions by cytokines tumor necrosis factor-alpha plus interferon-gamma Does not reduce leukocyte transmigration under flow. Am J Pathol. 2001;159:2281–2291. doi: 10.1016/s0002-9440(10)63078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin TA, Watkins G, Mansel RE, Jiang WG. Hepatocyte growth factor disrupts tight junctions in human breast cancer cells. Cell BiolInt. 2004;28:361–371. doi: 10.1016/j.cellbi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Mariscal L, Lechuga S, Garay E. Role of tight junctions in cell proliferation and cancer. Prog Histochem Cytochem. 2007;42:1–57. doi: 10.1016/j.proghi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Sommers CL, Gelmann EP, Kemler R, Cowin P, Byers SW. Alterations in beta-cateninphosphorylation and plakoglobin expression in human breast cancer cells. Cancer Res. 1994;54:3544–3552. [PubMed] [Google Scholar]
- 32.Sommers CL, Thompson EW, Torri JA, Kemler R, Gelmann EP, Byers SW. Cell adhesion molecule uvomorulin expression in human breast cancer cell lines: relationship to morphology and invasive capacities. Cell Growth Differ. 1991;2:365–372. [PubMed] [Google Scholar]
- 33.Nieman MT, Kim JB, Johnson KR, Wheelock MJ. Mechanism of extracellular domain-deleted dominant negative cadherins. J Cell Sci. 1999;112:1621–1632. doi: 10.1242/jcs.112.10.1621. [DOI] [PubMed] [Google Scholar]
- 34.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bazzoni G, Martinez-Estrada OM, Mueller F, Nelboeck P, Schmid G, Bartfai T, et al. Homophilic interaction of Junctional Adhesion Molecule. J Biol Chem. 2000 doi: 10.1074/jbc.M003946200. [DOI] [PubMed] [Google Scholar]
- 36.Ostermann G, Weber KS, Zernecke A, Schroder A, Weber C. JAM-1 is a ligand of the beta2 integrin LFA-1 involved in transendothelial migration of leukocytes. Nat Immunol. 2002;14:14. doi: 10.1038/ni755. [DOI] [PubMed] [Google Scholar]
- 37.Bazzoni G. The JAM family of junctional adhesion molecules. Curr Opin Cell Biol. 2003;15:525–530. doi: 10.1016/s0955-0674(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 38.Severson EA, Jiang L, Ivanov AI, Mandell KJ, Nusrat A, Parkos CA. Cis-dimerization mediates function of junctional adhesion molecule a. Mol Biol Cell. 2008;19:1862–1872. doi: 10.1091/mbc.E07-09-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]