Abstract
Background
Experimental and epidemiologic studies have suggested that high calcium intake is associated with decreased colon cancer risk. Yet very limited data are available for candidate genes in the calcium/vitamin D pathway and colon cancer risk. To address this, we evaluated whether calcium sensing receptor(CASR) single nucleotide polymorphisms(SNP) are associated with colon cancer risk. We also examined interactions between CASR and calcium and vitamin D intake, and previously genotyped vitamin D-related genes.
Methods
We conducted a large multi-center population-based case-control study of 1,600 cases and 1,949 controls. Seventeen tagging SNPs for CASR were selected from common SNPs (minor allele frequency ≥5%) based on resequencing data. Haplotypes were estimated and evaluated using HaploStats.
Results
We did not observe an association between any CASR genotypes or haplotypes and colon cancer risk overall. However, when stratified by anatomic site, statistically significant associations were seen with risk of proximal colon cancer (rs10934578 TT: OR 1.35, 95%CI 1.01-1.81; rs12485716 AG/AA: OR 0.84, 95%CI 0.71-1.00; rs4678174 CT/CC: OR 0.83, 95%CI 0.70-0.98; rs2270916 CC: OR 0.43, 95%CI 0.19-0.97). Concordantly, we observed a suggested association for a CASR haplotype (rs4678174, rs2270916) with risk of proximal colon cancer (global p-value=0.08). We did not observe any meaningful gene-environment (calcium and vitamin D) or gene-gene (CYP24A1, CYP27B1, and VDR) interactions with CASR genotypes and colon cancer risk.
Conclusion
Our study does not provide evidence for an overall association between CASR SNPs and colon cancer; however, results suggest a possible role of CASR on proximal colon cancer and subsite differences are consistent with known calcium biology. Nonetheless, these findings require confirmation.
Introduction
Substantial evidence from both epidemiologic (1-7) and experimental studies (8-12) support the role of dietary calcium as a protective factor in colon or colorectal cancer development(13). Similar findings were demonstrated in both a recent meta-analysis for milk and dairy products, the major sources of calcium in Western countries (14), and a pooled analysis evaluating milk and calcium consumption (15). In addition, three randomized trials found a reduction in colorectal adenoma recurrence with calcium supplementation (1200-2000 mg/day for three to four years) (16-19), an effect which persisted up to five years after the end of treatment(20). However, results from the Women’s Health Initiative, the only trial of calcium supplementation (1000 mg/day for seven years) with colorectal cancer as an endpoint, did not support the findings from observational studies (21) and trials of adenoma recurrence; but high calcium intake among participants at enrollment and a high proportion of calcium supplement use in the placebo group complicate the interpretation of this trial (22, 23).
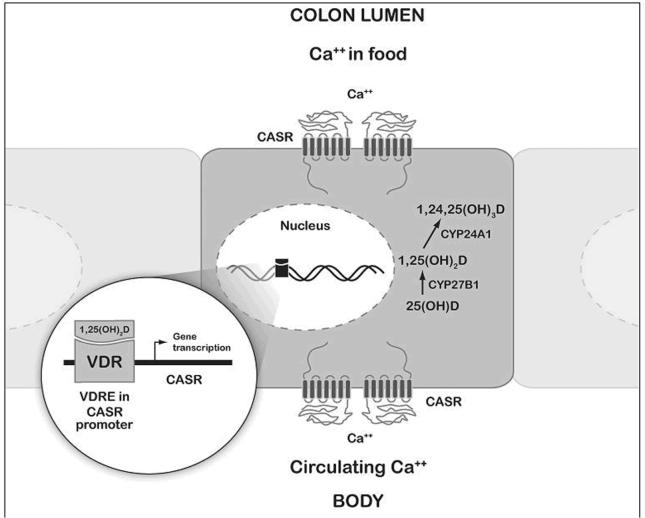
Evidence from experimental studies suggests that calcium prevents colon cancer development through two potentially relevant mechanisms: indirectly by binding free fatty acids and bile acids, and directly by inducing apoptosis and differentiation through binding to the calcium sensing receptor (CASR) (24). In addition to the important role of vitamin D in maintaining calcium homeostasis, the discovery of vitamin D-response elements in the promoter regions of CASR provides an intriguing link between calcium and vitamin D at the molecular level (Figure 1) (25). The primary focus thus far in the calcium/vitamin D pathway has been on polymorphisms in the vitamin D receptor (VDR); however, CASR, also expressed in the gastrointestinal tract, is an important candidate gene in this pathway that may prove helpful in elucidating mechanistic pathways through which calcium acts.
Figure 1. Calcium and vitamin D interrelated pathway in colon epithelial cell.
Circulating calcium and calcium in the colon lumen binds to CASR to activate signaling pathways involved in cell growth control and differentiation. Active vitamin D (1,25(OH)2) binds to the nuclear receptor VDR, which directly regulates the transcription of CASR through binding to two vitamin D response elements (VDRE) in the CASR promoter regions. 25-hydroxyvitamin D (25(OH)D) can be activated within the colon cell to 1,25(OH)2 by 1-α hydroxylase (CYP27B1) and deactivated by 24-α hydroxylase (CYP24A1).
The few studies (26-28) that have investigated the association between CASR and colorectal tumors have focused primarily on three non-synonymous SNPs, potentially overlooking the additional genetic variation in this large gene (102 kb) that is not sufficiently captured by these three SNPs; hence the need to take advantage of the recent genetic characterization of CASR to explore functional effects associated with its genetic variation (29). In our previous study, we observed no association between any specific genotype of these three SNPs and colorectal adenoma risk but a significant association with CASR diplotypes, suggesting that another polymorphism may be linked with these diplotypes (26). We comprehensively assessed the genetic variation of CASR in relation to colon cancer risk in a population-based case-control study of individuals from California, Minnesota, and Utah. We also examined whether CASR variants modified associations between colon cancer and calcium and vitamin D intake, and sun exposure, a marker for endogenous vitamin D production. As calcium and vitamin D are strongly interrelated at the physiological level, we explored gene-gene interaction between CASR variants with previously genotyped variants in vitamin D-related genes, including VDR, CYP24A1 (the vitamin D deactivating enzyme 24-alpha hydroxylase) and CYP27B1 (the vitamin D activating enzyme 1-alpha hydroxylase).
Materials and Methods
Study Population
Colon cancer cases and controls were recruited from: the Kaiser Permanente Medical Care Program (KPMCP) of Northern California; an eight-county area in Utah; and the metropolitan Twin Cities area of Minnesota. This multi-center population-based case-control study included 1,993 cases and 2,410 controls. Cases were aged 30 to 79 years at time of diagnosis of first primary colon cancer (ICD-O Ed.2. codes 18.0, 18.2-18.9) between October 1991 and September 1994. Cases with tumors in the rectosigmoid junction or rectum and cases with pathology report indicating Familial Adenomatous Polyposis, Crohn’s disease, or ulcerative colitis were not eligible. Incident cases of colon cancer were identified using a rapid-reporting system, with a majority interviewed within 4 months of diagnosis. Of the cases that were asked to participate in the study, 76% cooperated; which represents 65% of the eligible cases (for further details see (30)).
Controls were matched to cases by 5-year age groups and sex. Controls from KPMCP were randomly selected from membership lists. In Utah, controls who were under 65 years of age were randomly selected from lists generated using random-digit dialing and driver license lists, and those 65 years of age and older were randomly selected from Health Care Financing Administration lists. In Minnesota, control participants were identified from driver’s license or state identification lists. Of all controls asked to participate, 64% cooperated (30).
Tagging single nucleotide polymorphism (tagSNP) selection
TagSNP selection was based on existing resequencing data from SeattleSNPs (http://pga.gs.washington.edu/), which was limited to 23 HapMap CEPH individuals with European ancestry, given that White was the predominant racial group in our study population. The promoter region (2 kb upstream of Exon 1), complete coding sequence (7 exons, including intron/exon boundaries), conserved intronic regions, and a portion of the untranslated region (2 kb downstream of STP) of CASR was resequenced. Linkage disequilibrium (LD) structure of CASR was characterized by four blocks, encompassing areas in intron 1, 3, 4, and intron 5-exon6-intron 6 (the small block in intron 4 is not described as we only successfully genotyped one SNP within it). To include potentially functionally relevant SNPs, we selected all non-synonymous SNPs. Thereafter, we identified tagSNPs using the htSNP program developed by Clayton (31) to capture the common variation in the gene (minimum r2=0.80, minor allele frequency >5%, all non-synonymous SNPs were predefined as tagSNPs). Finally, we examined how well the selected tagSNPs were able to identify common haplotypes (>5%). To preserve a parsimonious set of tagSNPs, additional SNPs required for capturing haplotypes were tested to determine whether they could replace previously selected SNPs, yet still maintain a minimum r2 of 0.80. A total of 22 SNPs were selected for genotyping in CASR.
Genotyping
Of the 4,403 cases and controls with valid study data, 3680 (83% of cases and 85% of controls) provided a blood sample. Genomic DNA was extracted from peripheral blood lymphocytes or immortalized cell lines and was available from 1,600 cases (80% of cases) and 1,949 controls (81% of controls). Staff was blinded to case/control status and duplicate quality control samples, which were interspersed among plates (147 duplicates from controls). All genotyping was performed by MALDITOF mass spectrometry on the Sequenom MassARRAY7K platform using the iPLEX Gold (low-plex) reaction. In total, we received genotyping results for 17 CASR SNPs.
The call rate was >98% for all 17 SNPs. Blinded duplicate samples displayed >98% concordance for any SNP. Using a goodness-of-fit test, the allele frequencies among Caucasian controls were consistent with Hardy-Weinberg Equilibrium (p >0.01) for all SNPs. The 17 successfully genotyped tagSNPs covered the genetic variation of CASR with a minimum r2 of 0.38 and a mean r2 of 0.97.
Dietary and lifestyle data
Detailed in-person interviews were used to collect demographic, dietary and lifestyle data from all eligible study participants. Study participants were asked about their lifestyle during the year 2 years prior to the date of diagnosis (cases) or selection (controls). During the in-person interview, information was collected on dietary intake, physical activity, medical history and drug use, demographic factors, smoking, reproductive history (for women) and family history of cancer and colorectal polyps. Quality control methods have been described in detail elsewhere (32).
Dietary intake was ascertained using a diet-history questionnaire, designed and validated for the Coronary Artery Risk Development in Young Adults (CARDIA) study (33, 34). Participants were asked to recall foods eaten, the frequency they were eaten, serving size, and use of fats during food preparation. The nutrient database was based on the University of Minnesota Nutrient Data System for Research. In a comparative validity study, assessment of dietary calcium intake was measured well (Pearson correlations of 0.79 and 0.70 for men and women)(35). Multivitamin, calcium and vitamin D supplement use were also ascertained. We defined supplemental vitamin D as 10 μg(400IU)/day if a participant reported either regular multivitamin or vitamin D supplement use (three times/week for at least one month). Total vitamin D combines dietary and supplemental vitamin D intake. Participants were asked if they used anti-acid medications (Rolaids or Tums); however, calcium was restricted to dietary intake only because supplemental dosage was not provided and doses in multivitamins are often low and highly variable.
UV-weighted hours of sun exposure (UV index-hours/week) was based on the average hours per week spent outdoors in the daylight reported by subjects during each season (spring, summer, fall, winter) of the referent year, multiplied by the UV index for each season in the geographic area of the study center, and divided by four to average over the four seasons (36). Height and weight measurements were used to determine body mass index (BMI) at referent year. Physical activity was assessed using a questionnaire adapted from the CARDIA study to include more details on amount of time and frequency of activities over a 20 year time period (37, 38). Regular NSAID use was defined as at least three times a week for at least one month.
Statistical analyses
Unconditional logistic regression, adjusted for age, sex, race and study center, was used to estimate odds ratios (OR) and 95% confidence intervals (95% CI) for the association between individual polymorphisms and colon cancer. Dietary factors included as potential confounders or effect modifiers were energy adjusted using the residual method (39). The distribution of intake among controls was used to determine cutpoints. Additional adjustment for education, income, BMI, cigarette smoking, physical activity, long-term alcohol use, NSAID use, family history of colorectal cancer, dietary fiber, folate, red meat, fat intake, and multivitamin use did not result in meaningful changes of risk estimates and were not included in the analyses. Global tests of association were conducted by simultaneously including genotypes (heterozygotes and homozygotes rare allele) of all CASR SNPs in a model and comparing it to a model that included none of the genotypes. Adjustment for multiple testing was achieved through this multi-locus global test (degrees of freedom = 2× number of SNPs in a gene) within the likelihood ratio chi-square test. When SNPs were highly correlated (r2>0.80), only one of the SNPs were included in the gene-based model. All analyses were conducted using SAS version 9.1 (SAS Institute, Cary, NC).
Haploview was used to examine measures of LD (D’ and r2) between SNPs and to define haplotype-block structure based on Gabriel’s definition (40). Haplotype frequencies were estimated and associations evaluated using HaploStats (Version 1.3.1) in R (version 2.4.1), assuming an additive model unless otherwise indicated. The most common haplotype was used as the reference group and rare haplotypes (frequency <5%) were combined into a single category. Analyses for haplotypes were restricted to non-Hispanic whites, the predominant racial group in our study, and haplotype frequencies and associations differ by racial group. All haplotype analyses were adjusted for age, sex, and study center. A global score statistic was used to evaluate the overall difference in haplotype frequencies between cases and controls.
Polytomous regression was used to estimate associations between genotypes and risk of proximal colon cancer (cecum, ascending colon, hepatic flexure and transverse colon) and distal colon cancer (splenic flexure, descending colon, and sigmoid colon).
To evaluate whether the presence of a genotype or haplotype modified the association between calcium and vitamin D intake (as well as sun exposure) and colon cancer risk and whether there was evidence of a gene-gene interaction between CASR, CYP24A1, CYP27B1 and VDR, we investigated interactions through the inclusion of cross-product terms in the regression models. We performed an omnibus test for interaction between CASR with the variable of interest (e.g. calcium) by simultaneously including all cross-product terms for the CASR genotypes (coded as dummy variables for heterozygotes and variant homozygotes) with the variable of interest in a model (coded as continuous for dietary factors or UV-weighted sun exposure) and comparing that to a model including only the main effects for genotypes and the variable of interest. In omnibus tests for interaction between two genes, genotypes for both genes were reduced to a binary variable combining heterozygotes and variant homozygotes. If the omnibus test for interaction was statistically significant, we further investigated multiplicative interaction between individual SNPs and the variable of interest, using the log likelihood ratio test to compare the fit of models with and without interaction terms.
Several publicly available web-based tools were used to assess the potential functional significance of variants, including: the UCSC Genome Browser to obtain conservation scores (41), SIFT (42) and PolyPhen (43) to evaluate the functional effect of non-synonymous SNPs, the Splice Site Prediction tool by Neural Network (44) to identify potential splice sites, ESEfinder (45) and RESCUEESE (46) to evaluate whether a SNP was located in a potential exonic splice enhancer, and UTRScan to identify areas within the untranslated region (UTR) that may be associated with mRNA stability and translation (47).
Results
The majority of the study population was non-Hispanic whites, with a slightly higher proportion among controls (Table 1). Controls were more likely to be female, be college graduates and have a higher income. Dietary intake of calcium and vitamin D were higher among controls than cases. Multivitamin use and use of calcium and vitamin D supplements was slightly higher in controls. An inverse association between the highest quartile of dietary calcium and colon cancer was observed (proximal: OR=0.72; 95% CI, 0.56-0.91; distal: OR= 0.63; 95% CI, 0.49-0.80), but nonsignificant inverse associations for the highest quartiles of dietary vitamin D (proximal: OR=0.83; 95% CI, 0.66-1.05; distal: OR=0.80; 95% CI, 0.63-1.01) and sunshine exposure (proximal: OR = 0.71; 95% CI, 0.55-0.92; distal: OR=0.84; 95% CI, 0.65-1.09). These results have been previously published (7).
Table 1.
Characteristics of cases and controls
| Cases (n=1600) | Controls (n=1949) | |
|---|---|---|
| Age (years)* | 64.9 ± 9.8 | 65.0 ± 10.1 |
| Sex | ||
| Male | 895 (55.9) | 1036 (53.2) |
| Female | 705 (44.1) | 913 (46.8) |
| Race | ||
| White, not Hispanic | 1461 (91.4) | 1814 (93.1) |
| Hispanic | 62 (3.9) | 78 (4.0) |
| African American | 72 (4.5) | 53 (2.7) |
| Other | 4 (0.2) | 3 (0.2) |
| Study center | ||
| Kaiser | 769 (48.1) | 804 (41.3) |
| Utah | 578 (36.1) | 796 (40.8) |
| Minnesota | 253 (15.8) | 349 (17.9) |
| Education | ||
| Less than 12 years | 262 (16.4) | 249 (12.8) |
| High school graduate | 451 (28.2) | 545 (28.0) |
| Some college or post-high school | 530 (33.1) | 634 (32.6) |
| College graduate or higher | 357 (22.3) | 520 (26.7) |
| Income | ||
| <$20,000 | 401 (27.0) | 455 (24.8) |
| <20,000-40,000 | 533 (35.9) | 627 (34.2) |
| <40,000-<60,000 | 348 (23.4) | 438 (23.9) |
| <60,000 + | 203 (13.7) | 313 (17.1) |
| Mean UV-weighted hours of sun | ||
| 85.8 ± 70.9 | 87.4 ± 68.7 | |
| exposure (UV index hours/week) | ||
| Mean dietary calcium (mg/day)† | 1001.3 ± 392.2 | 1058.9 ± 411.6 |
| Mean total vitamin D (μg/day)† | 10.6 ± 6.0 | 11.1 ± 6.2 |
| Multivitamin supplement use‡ | 520 (32.5) | 654 (33.6) |
| Calcium supplement use‡ | 194 (12.1) | 316 (16.2) |
| Vitamin D supplement use‡ | 25 (1.6) | 53 (2.7) |
| Tumor site | ||
| Distal | 790 (49.4) | - |
| Proximal | 771 (48.2) | - |
| Unknown | 39 (2.4) | - |
Continuous variables are displayed as mean values ± standard deviation and frequencies are displayed as counts (percentage).
Defined as age at diagnosis for cases and age at recruitment for controls.
Energy adjusted (residual method) dietary intake.
Supplement use defined as regular use (at least three times a week for at least one month) over the referent period.
Overall, we did not observe a statistically significant association between any of the CASR polymorphisms and colon cancer risk (Table 2). The global association p-value for SNPs in CASR and colon cancer was 0.52. Because the association with colon cancer differs for several risk factors by anatomic site, we examined associations for distal and proximal colon cancer. Four CASR variants displayed a statistically significant association with risk of proximal colon cancer (Table 2); the global p-value was low but not significant (p=0.10; df=32). We observed a positive association between a CASR variant, IVS3+1048, and risk of proximal colon cancer (all ethnicities: OR TT vs. GG = 1.35; 95% CI, 1.01-1.81; non-Hispanic whites: OR = 1.38; 95% CI, 1.02-1.85). CASR variant IVS3-685 was associated with a decreased risk of proximal cancer (all ethnicities: OR AA+AG vs. GG =0.84; 95% CI, 0.71-1.00; non-Hispanic whites: OR = 0.81; 95% CI, 0.68-0.97) for carriers of at least one copy of the rare allele. A statistically significant inverse association with proximal colon cancer risk was found for IVS5-90 (all ethnicities: OR CC+CT vs. TT = 0.83; 95% CI, 0.70-0.98; non-Hispanic whites: OR = 0.80; 95% CI, 0.67-0.95). Furthermore, we observed an inverse association for IVS6+16 and proximal colon cancer risk among all ethnicities: OR CC vs. TT = 0.43 (95% CI, 0.19-0.97); and among non-Hispanic whites: OR = 0.44 (95% CI, 0.20-1.00). The IVS3+1048 variant was in strong LD with IVS3-685, IVS5-90 and IVS6+16 (D’=1.0), but weakly correlated with these three variants (r2=0.11, r2=0.12 and r2=0.07, respectively). IVS3-685 was highly correlated with IVS5-90 (r2=0.89) and moderated correlated with IVS6+16 (r2=0.59). The two variants IVS5-90 and IVS6+16 were in strong LD (D’=1.0) and moderately correlated (r2=0.53) with each other. The associations observed for variants IVS3+1048, IVS3-685, IVS5-90, and IVS6+16 were slightly stronger after adjustment for each other and all other SNPs in CASR (data not shown).
Table 2.
ORs and 95% CIs for the association between polymorphisms in CASR and colon cancer risk
| All colon cancer | Proximal colon cancer | Distal colon cancer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position/ Genotype | Controls N |
Cases N |
OR* | 95% CI | P for trend | Cases N |
OR* | 95%CI | P for trend | Cases N |
OR* | 95%CI | P for trend |
|
-465T>C (rs6678267) |
|||||||||||||
| CC | 1270 | 1039 | 1.00 | - | 505 | 1.00 | - | 508 | 1.00 | - | |||
| CT | 563 | 485 | 1.07 | 0.93-1.24 | 232 | 1.05 | 0.87-1.26 | 242 | 1.10 | 0.91-1.32 | |||
| TT | 87 | 56 | 0.80 | 0.57-1.13 | 0.93 | 26 | 0.75 | 0.48-1.18 | 0.69 | 28 | 0.83 | 0.54-1.29 | 0.82 |
| TT + CT vs. CC | 1.04 | 0.90-1.19 | 1.01 | 0.84-1.21 | 1.06 | 0.89-1.27 | |||||||
|
IVS1+2998A>G (rs672782) |
|||||||||||||
| GG | 778 | 643 | 1.00 | - | 311 | 1.00 | - | 315 | 1.00 | - | |||
| GA | 869 | 713 | 1.01 | 0.87-1.17 | 337 | 0.98 | 0.82-1.18 | 360 | 1.05 | 0.88-1.26 | |||
| AA | 272 | 219 | 0.98 | 0.80-1.21 | 0.93 | 110 | 1.02 | 0.79-1.32 | 0.97 | 103 | 0.94 | 0.72-1.23 | 0.88 |
| AA + GA vs. GG | 1.00 | 0.88-1.15 | 0.99 | 0.84-1.18 | 1.02 | 0.86-1.21 | |||||||
|
IVS1+12198G>A (rs6802749) |
|||||||||||||
| GG | 1329 | 1097 | 1.00 | - | 517 | 1.00 | - | 554 | 1.00 | - | |||
| GA | 544 | 444 | 1.00 | 0.86-1.16 | 224 | 1.07 | 0.88-1.28 | 207 | 0.93 | 0.77-1.12 | |||
| AA | 62 | 42 | 0.81 | 0.54-1.21 | 0.55 | 20 | 0.83 | 0.49-1.38 | 0.91 | 22 | 0.83 | 0.50-1.37 | 0.31 |
| AA + GA vs. GG | 0.98 | 0.85-1.13 | 1.04 | 0.87-1.25 | 0.92 | 0.76-1.10 | |||||||
|
IVS1+25086G>A (rs17203502) |
|||||||||||||
| GG | 526 | 431 | 1.00 | - | 218 | 1.00 | - | 204 | 1.00 | - | |||
| GA | 953 | 780 | 0.98 | 0.84-1.15 | 370 | 0.93 | 0.76-1.13 | 388 | 1.03 | 0.84-1.26 | |||
| AA | 461 | 382 | 0.99 | 0.82-1.20 | 0.92 | 180 | 0.93 | 0.73-1.17 | 0.51 | 194 | 1.05 | 0.83-1.33 | 0.68 |
| AA + GA vs. GG | 0.99 | 0.85-1.15 | 0.93 | 0.77-1.12 | 1.03 | 0.86-1.25 | |||||||
|
IVS1-18718C>A (rs1354162) |
|||||||||||||
| CC | 1530 | 1283 | 1.00 | - | 625 | 1.00 | - | 629 | 1.00 | - | |||
| CA | 338 | 267 | 0.95 | 0.79-1.13 | 122 | 0.89 | 0.71-1.12 | 137 | 0.99 | 0.79-1.23 | |||
| AA | 29 | 17 | 0.71 | 0.39-1.29 | 0.29 | 8 | 0.67 | 0.31-1.48 | 0.18 | 8 | 0.67 | 0.31-1.49 | 0.57 |
| AA + CA vs. CC | 0.93 | 0.78-1.10 | 0.87 | 0.70-1.09 | 0.96 | 0.78-1.20 | |||||||
|
IVS1-4705C>T (rs9875101) |
|||||||||||||
| CC | 957 | 784 | 1.00 | - | 371 | 1.00 | - | 395 | 1.00 | - | |||
| CT | 804 | 663 | 1.00 | 0.87-1.15 | 314 | 1.01 | 0.84-1.20 | 332 | 0.99 | 0.83-1.18 | |||
| TT | 178 | 143 | 0.97 | 0.77-1.24 | 0.89 | 82 | 1.19 | 0.89-1.59 | 0.37 | 57 | 0.77 | 0.56-1.06 | 0.25 |
| TT + CT vs. CC | 1.00 | 0.87-1.14 | 1.04 | 0.88-1.23 | 0.95 | 0.81-1.13 | |||||||
|
IVS1-160T>C (rs34178491) |
|||||||||||||
| TT | 1339 | 1121 | 1.00 | - | 535 | 1.00 | - | 560 | 1.00 | - | |||
| CT | 515 | 416 | 0.97 | 0.83-1.13 | 206 | 1.01 | 0.83-1.22 | 198 | 0.92 | 0.76-1.12 | |||
| CC | 58 | 40 | 0.83 | 0.55-1.25 | 0.42 | 21 | 0.91 | 0.55-1.52 | 0.88 | 19 | 0.79 | 0.47-1.35 | 0.27 |
| CC + CT vs. TT | 0.96 | 0.82-1.11 | 1.00 | 0.83-1.20 | 0.91 | 0.76-1.10 | |||||||
|
IVS3+298A>T rs3749204 |
|||||||||||||
| AA | 482 | 419 | 1.00 | - | 214 | 1.00 | - | 197 | 1.00 | - | |||
| AT | 930 | 765 | 0.97 | 0.82-1.14 | 354 | 0.87 | 0.71-1.07 | 393 | 1.06 | 0.87-1.31 | |||
| TT | 472 | 373 | 0.93 | 0.77-1.13 | 0.50 | 181 | 0.88 | 0.70-1.12 | 0.29 | 181 | 0.96 | 0.76-1.23 | 0.77 |
| TT + AT vs. AA | 0.96 | 0.82-1.11 | 0.88 | 0.72-1.06 | 1.03 | 0.85-1.25 | |||||||
|
IVS3+1048G>T (rs10934578) |
|||||||||||||
| GG | 907 | 731 | 1.00 | - | 344 | 1.00 | - | 365 | 1.00 | - | |||
| GT | 864 | 711 | 1.04 | 0.90-1.19 | 337 | 1.04 | 0.87-1.25 | 361 | 1.06 | 0.89-1.26 | |||
| TT | 169 | 152 | 1.13 | 0.89-1.44 | 0.33 | 86 | 1.35 | 1.01-1.81 | 0.09 | 62 | 0.93 | 0.67-1.27 | 0.98 |
| TT + GT vs. GG | 1.05 | 0.92-1.20 | 1.09 | 0.92-1.29 | 1.04 | 0.88-1.23 | |||||||
|
IVS3-685G>A (rs12485716) |
|||||||||||||
| GG | 955 | 799 | 1.00 | - | 411 | 1.00 | - | 372 | 1.00 | - | |||
| AG | 784 | 630 | 0.95 | 0.82-1.09 | 293 | 0.86 | 0.72-1.03 | 321 | 1.04 | 0.87-1.24 | |||
| AA | 173 | 148 | 0.98 | 0.77-1.25 | 0.61 | 58 | 0.75 | 0.54-1.04 | 0.03 | 85 | 1.20 | 0.90-1.61 | 0.27 |
| AA + AG vs. GG | 0.95 | 0.83-1.09 | 0.84 | 0.71-1.00 | 1.07 | 0.90-1.26 | |||||||
|
IVS4+2761C>T (rs35387386) |
|||||||||||||
| CC | 1823 | 1516 | 1.00 | - | 730 | 1.00 | - | 747 | 1.00 | - | |||
| CT | 104 | 64 | 0.73 | 0.53-1.00 | 30 | 0.71 | 0.47-1.08 | 34 | 0.78 | 0.52-1.16 | |||
| TT† | 1 | 1 | - | - | 0.06 | 1 | - | - | - | 0 | - | - | 0.18 |
| TT + CT vs. CC | 0.73 | 0.53-1.00 | 0.73 | 0.48-1.10 | 0.77 | 0.52-1.14 | |||||||
|
IVS4-1185C>T (rs35780274) |
|||||||||||||
| CC | 628 | 535 | 1.00 | - | 257 | 1.00 | - | 267 | 1.00 | - | |||
| CT | 917 | 732 | 0.96 | 0.82-1.12 | 340 | 0.92 | 0.76-1.12 | 369 | 0.97 | 0.80-1.17 | |||
| TT | 365 | 303 | 1.01 | 0.83-1.22 | 0.96 | 161 | 1.10 | 0.87-1.40 | 0.57 | 138 | 0.93 | 0.72-1.18 | 0.54 |
| TT + CT vs. CC | 0.97 | 0.84-1.12 | 0.97 | 0.81-1.17 | 0.96 | 0.80-1.15 | |||||||
|
IVS5-90C>T (rs4678174) |
|||||||||||||
| TT | 840 | 708 | 1.00 | - | 363 | 1.00 | - | 332 | 1.00 | - | |||
| CT | 865 | 670 | 0.90 | 0.78-1.04 | 314 | 0.83 | 0.69-0.99 | 335 | 0.97 | 0.81-1.16 | |||
| CC | 239 | 214 | 1.00 | 0.81-1.25 | 0.57 | 90 | 0.83 | 0.62-1.09 | 0.05 | 119 | 1.19 | 0.91-1.54 | 0.41 |
| CC + CT vs. TT | 0.92 | 0.81-1.06 | 0.83 | 0.70-0.98 | 1.01 | 0.85-1.20 | |||||||
|
IVS6+16T>C (rs2270916) |
|||||||||||||
| TT | 1425 | 1202 | 1.00 | - | 593 | 1.00 | - | 584 | 1.00 | - | |||
| CT | 476 | 365 | 0.94 | 0.80-1.10 | 169 | 0.87 | 0.71-1.07 | 183 | 0.97 | 0.80-1.19 | |||
| CC | 40 | 28 | 0.85 | 0.52-1.39 | 0.32 | 7 | 0.43 | 0.19-0.97 | 0.04 | 20 | 1.27 | 0.73-2.19 | 0.82 |
| CC + CT vs. TT | 0.93 | 0.80-1.08 | 0.84 | 0.69-1.02 | 1.00 | 0.82-1.21 | |||||||
|
Ex7+1224G>T A986S (rs1801725) |
|||||||||||||
| GG | 1476 | 1197 | 1.00 | - | 572 | 1.00 | - | 594 | 1.00 | - | |||
| GT | 430 | 371 | 1.08 | 0.92-1.26 | 182 | 1.10 | 0.90-1.34 | 181 | 1.06 | 0.87-1.30 | |||
| TT | 33 | 27 | 1.00 | 0.60-1.67 | 0.45 | 14 | 1.08 | 0.58-2.04 | 0.37 | 13 | 0.95 | 0.50-1.83 | 0.68 |
| TT + GT vs. GG | 1.07 | 0.92-1.25 | 1.10 | 0.91-1.33 | 1.05 | 0.87-1.28 | |||||||
|
Ex7+1236A>G R990G (rs1042636) |
|||||||||||||
| AA | 1640 | 1347 | 1.00 | - | 646 | 1.00 | - | 670 | 1.00 | - | |||
| GA | 262 | 224 | 1.04 | 0.86-1.27 | 112 | 1.09 | 0.85-1.38 | 106 | 1.00 | 0.78-1.27 | |||
| GG | 12 | 7 | 0.72 | 0.28-1.84 | 0.88 | 5 | 1.06 | 0.37-3.03 | 0.51 | 2 | 0.41 | 0.09-1.86 | 0.65 |
| GG + GA vs. AA | 1.03 | 0.85-1.25 | 1.09 | 0.86-1.37 | 0.97 | 0.76-1.24 | |||||||
|
Ex7+1299G>C E1011Q (rs1801726) |
|||||||||||||
| CC | 1725 | 1425 | 1.00 | - | 684 | 1.00 | - | 707 | 1.00 | - | |||
| CG | 180 | 149 | 0.97 | 0.77-1.22 | 77 | 1.06 | 0.80-1.41 | 68 | 0.88 | 0.66-1.19 | |||
| GG | 5 | 4 | 0.74 | 0.19-2.84 | 0.71 | 2 | 0.79 | 0.15-4.23 | 0.77 | 2 | 0.72 | 0.14-3.86 | 0.38 |
| GG + CG vs. CC | 0.97 | 0.77-1.21 | 1.05 | 0.80-1.40 | 0.88 | 0.66-1.18 | |||||||
| Global P | 0.52 | 0.10 | 0.93 | ||||||||||
Adjusted for age, race, sex and study center.
Only 1 case and control with TT genotype, thus estimates are not provided.
Haplotype analysis did not reveal any statistically significant associations between CASR haplotypes and risk of colon cancer overall; however, an association with proximal colon cancer was observed (Table 3). The haplotype carrying the rare allele of IVS3+1048 in Block 2 (A-T) displayed a positive but statistically non-significant association with proximal colon cancer risk. This weaker association for haplotype compared with genotype is to be expected because the haplotype analysis is conducted under an additive model while the genotype analysis showed statistically significant associations under a recessive model for IVS3+1048. As with the genotype analyses for proximal colon cancer, we observed a significant association for a haplotype in Block 3 of CASR, containing IVS5-90 and IVS6+16 (p-value for global test =0.08; Table 3). We observed an inverse association between the C-C haplotype and proximal colon cancer risk, with an OR of 0.80 (95% CI, 0.67-0.97).
Table 3.
Association between CASR haplotypes and Colon Cancer among non-Hispanic whites
| All colon cancer | Proximal colon cancer | Distal colon cancer | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Controls % | Cases % | OR*(95% CI) | p | Cases % | OR*(95% CI) | p | Cases % | OR*(95% CI) | p |
| Block 1(IVS1+12198G>A, IVS1+25086G>A, IVS1-18718C>A)† | ||||||||||
| G-A-C | 37.8 | 37.7 | 1.00 | 37.2 | 1.00 | 38.4 | 1.00 | |||
| G-G-C | 34.7 | 35.2 | 1.02 (0.91-1.14) | 35.5 | 1.04 (0.90-1.21) | 34.8 | 0.99 (0.86-1.15) | |||
| A-G-C | 16.3 | 16.3 | 1.00 (0.87-1.16) | 17.0 | 1.06 (0.88-1.27) | 15.6 | 0.94 (0.79-1.14) | |||
| G-A-A | 10.3 | 10.0 | 0.96 (0.81-1.15) | 9.3 | 0.91 (0.73-1.14) | 10.4 | 0.99 (0.80-1.23) | |||
| Global P | 0.64 | 0.83 | 0.97 | |||||||
| Block 2(IVS3+298A>T, IVS3+1048G>T)‡ | ||||||||||
| T-G | 50.3 | 49.6 | 1.00 | 48.7 | 1.00 | 50.2 | 1.00 | |||
| A-T | 31.2 | 32.2 | 1.04 (0.93-1.16) | 33.7 | 1.11 (0.97-1.28) | 31.0 | 0.98 (0.85-1.14) | |||
| A-G | 18.4 | 18.1 | 1.00 (0.87-1.14) | 17.5 | 0.98 (0.83-1.16) | 18.6 | 1.01 (0.85-1.19) | |||
| Global P | 0.89 | 0.42 | 0.97 | |||||||
| Block 3(IVS5-90T>C, IVS6+16T>C)§ | ||||||||||
| T-T | 66.6 | 67.7 | 1.00 | 70.1 | 1.00 | 65.8 | 1.00 | |||
| C-T | 18.6 | 18.4 | 0.98 (0.86-1.11) | 17.5 | 0.90 (0.76-1.05) | 19.3 | 1.05 (0.89-1.23) | |||
| C-C | 14.7 | 13.8 | 0.93 (0.81-1.07) | 12.4 | 0.80 (0.67-0.97) | 14.9 | 1.04 (0.87-1.23) | |||
| Global P | 0.48 | 0.08 | 0.79 | |||||||
Adjusted for age, sex, and study center.
Loci of SNPs included in block are in the following order: rs6802749, rs17203502, and rs1354162.
Loci of SNPs included in block are in the following order: rs3749204, rs10934578.
Loci of SNPs included in block are in the following order: rs4678174, rs2270916.
We did not observe any statistically significant interactions between genotypes in CASR and UV-weighted hours of sun exposure (global p-value =0.09; df =32), dietary calcium (global p-value =0.75; df =32) or total vitamin D (global p-value =0.59; df =32) with overall colon cancer risk or by cancer subsite (data not shown) with one exception. An interaction between CASR genotypes and total vitamin D was observed (global p-value =0.05; df=31) with distal colon cancer risk, but further SNP by SNP investigation did not portray an obvious association. There was a suggestion of a decrease in risk of distal colon cancer associated with a change of 10 μg of total vitamin D among individuals with either none (OR =0.79; 95% CI, 0.65-0.97) or both copies (OR=0.46; 95% CI, 0.26-0.82) of the rare allele for IVS1-4705C>T; however, this relationship is difficult to explain from a biological perspective as no clear demonstration of risk is associated with the carrying of a specific allele. Associations observed for CASR haplotypes and colon cancer risk did not differ significantly by sunlight exposure, dietary calcium, or total vitamin D and were similar by cancer subsite (data not shown). There was no evidence of an interaction between genotypes in CASR and genotypes in VDR (Bsm1 and Fok1) with colon cancer (global p-value =0.39; df =32), proximal cancer (global p-value =0.88; df =32) and distal cancer risk (global p-value =0.37; df =32). Similarly, genotypes in CASR did not display a statistically significant interaction with CYP24A1 or CYP27B1 genotypes and risk of colon cancer overall or site-specific colon cancer (data not shown).
Discussion
We observed a statistically significant association between four genetic variants in CASR and proximal colon cancer risk. The global test of association between CASR and proximal colon cancer risk was low but not significant. We hypothesized that variants in CASR might modify the association between UV-weighted hours of sun exposure, calcium and vitamin D intake or genetic variants in vitamin D related genes on colon cancer risk as it is a key gene in the calcium/vitamin D pathway; however, there was no meaningful evidence of such interactions between CASR genotypes or haplotypes with any of these factors either overall or by anatomic site.
CASR is suggested to be a key component of the pathway through which calcium mediates its anti-carcinogenic effect on colorectal cancer development (48-53). CASR detects extracellular calcium and has a crucial role in calcium homeostasis. CASR is expressed throughout the entire gastrointestinal tract (54-58); in the colon, it is expressed on both the basolateral and luminal surfaces of colon cells, suggesting that CASR senses changes in calcium concentrations in the lumen of the colon as well as circulating concentrations (51, 56, 58). A recent review (51) of the molecular mechanisms of CASR in colorectal cancer indicated that signaling pathways involved in cell growth control and differentiation are activated by calcium through CASR, including promotion of E-cadherin expression, suppression of β-catenin/T cell factor (48) activation, and activation of the p38 mitogen-activated protein kinase (MAPK) cascade (59). Furthermore, studies have shown that the cell proliferative effect of low intestinal calcium concentrations is likely to be mediated by CASR via protein kinase C-signaling activation, leading to upregulation of c-myc expression (49, 50).
To our knowledge, this is the first study to investigate CASR variants and colon cancer using a comprehensive approach to capture common genetic variation; only the three non-synonymous variants (Ex7+1224(A986S), Ex7+1236(R990G), Ex7+1299(E1011Q)) we examined have been previously evaluated in relation to colorectal adenoma risk. Consistent with our results, that study found no association between these SNPs but rather a diplotype with distal colorectal adenoma risk (26). In our study, IVS3+1048 was associated with an increased risk of proximal colon cancer, but it is located within an intronic area that is not highly conserved (conservation score=0.01 (41)) and, thus, may not be responsible for the observed association. This variant is in LD (r2>0.5) with nine other CASR variants including nonsynonymous variant Ex7+1224 (A986S; D’=1.0 and r2=0.57) and a variant in the 3′ untranslated region (UTR) Ex7-1125delA (D’=1.0 and r2=0.89). We observed a consistent but statistically non-significant increased risk of proximal colon cancer with Ex7+1224 (A986S); however, this variant is suggested to have little functionality (SIFT= tolerated; PolyPhen= benign) despite being highly conserved (conservation score =1.0). A second linked variant (Ex7-1125delA) is a highly conserved (conservation score=0.99) deletion in the 3′ UTR but is also not predicted to be functionally important. A variant in the 3′ UTR, nonetheless, could be important as elements located in this region are involved in the stability and expression of mRNA (60); however, no such element was detected by UTRScan in the area immediately surrounding this variant. Hence, evidence supporting a specific variant as being responsible for the association with IVS3+1048 is lacking.
Three linked intronic CASR variants, IVS3-685, IVS5-90 and IVS6+16, were associated with a decreased risk of proximal colon cancer in our study. However, all three variants are not evolutionarily conserved (conservation score <0.1). IVS3-685, IVS5-90 and IVS6+16 are in strong LD with thirteen other CASR variants (r2:0.46-1.0). Only one of these variants, IVS4+2762, is correlated with all three tagSNPs (r2: 0.53-1.0) and highly conserved (conservation score =0.76), but the potential biologic importance of this intronic SNP is unknown. Although not residing in a highly conserved area, two variants in the 3′ UTR were also strongly correlated (Ex7-107: r2=0.90 and 0.80 with IVS5-90 and IVS3-685, respectively; and Ex7-212: r2=0.86 with IVS6+16). Accordingly, there are several potentially interesting candidates but as all three SNPs are in LD it is unclear whether one or all of these variants are responsible for the associations observed.
Growing evidence suggests that the effect of several risk factors may differ in the development of tumors within the proximal and distal colon; molecular and functional differences result in one site being more susceptible to specific exposures, such as diet (61-63). In particular, the prolonged presence in the proximal colon of bile acids produced by the liver and modified by the intestinal microflora could induce an increase in cell proliferation rates (63). Because calcium has the ability to bind and neutralize secondary bile acids and free fatty acids, the beneficial effect of calcium may be stronger in the proximal colon than the distal colon where, because of such binding as well as resorption, the bile acid concentrations are much lower. Results from our study suggest that calcium may also be mediating its effect in the proximal colon through CASR. Experimental studies have demonstrated that the majority of calcium absorption in the large intestine occurs in the proximal (cecum and ascending) colon (64-66). This is consistent with our findings of a statistically significant association between CASR variants and cancer in the proximal colon, where calcium may be more influential in reducing risk. However, results from epidemiologic studies have been mixed. Twelve observational studies have presented results on the effect of calcium by colon subsite (3-7, 67-73). Four of these studies (3-5, 67) found inverse associations for calcium to be stronger for proximal colon cancer than distal colon cancer, whereas six studies (6, 7, 68, 69, 71, 72) observed stronger associations for distal colon cancer, and two did not find a statistically significant association for either site (70, 73). In our study, dietary calcium was associated with a reduced risk of colon cancer in both sites, but this was slightly stronger for distal cancer (7). Further studies assessing the role of CASR variants are required to verify whether this association is, indeed, site-specific.
Our study has some strengths as well as limitations. As this gene has been investigated only to a limited extent, this study provides unique information on the role of CASR and its association with colon cancer risk. By basing our tagSNP selection on resequencing data, we were able to conduct a more comprehensive analysis of common genetic variation of CASR and colon cancer risk than the traditional candidate SNP approach. However, as CASR is a very large gene, the entire intronic region of CASR was not resequenced, making it probable that we will have missed some genetic variation (41% of gene sequenced). Nonetheless, the resequencing strategy by SeattleSNPs focused on regions of the gene that are likely to be most functionally relevant (promoter, coding regions, untranslated regions, and conserved intronic regions); thus, resequencing of additional areas is not likely to reveal additional putatively functional variants. Because of genotyping failure, our tagSNPs do not fully cover the entire genetic variation of CASR. However, our tagSNPs performed well by tagging 123 detected CASR variants with a mean r2 of 0.97. Of the 123 common variants detected through resequencing, only seven were tagged at an r2 between 0.38 to 0.71.
Other advantages of this study are the large number of cases and the detailed data collected on dietary habits and other environmental factors related to colon cancer, thus allowing us to control for important confounding factors. As this is a case-control study, dietary data were collected retrospectively and may be subject to recall bias; however, data were obtained rapidly after enrollment and collected with detailed and validated procedures (33, 34). A limitation of our data is that only dietary calcium intake was assessed. Inclusion of supplemental calcium from multivitamins or anti-acid medications, potentially important sources of calcium, would provide a better indicator of total calcium intake than diet alone. The average daily dietary calcium intake among this study population suggests a high calcium status (1056 mg), about 200 mg higher than the US national average (863 mg) (74); it is possible that specific CASR variants may be functionally relevant only among a subset of individuals with nutrient deficiency, which may explain the lack of interaction between CASR variants and calcium intake. This study had 80% power to detect a multiplicative interaction OR of 2.2, assuming a main-effects OR of 1.4 for the CASR genotype and an OR of 0.67 for calcium intake above the mean.
In summary, we observed a statistically significant association between CASR variants and proximal colon cancer risk but not distal colon cancer risk. Although this subset distinction is plausible, these results should be considered preliminary and additional studies are needed to confirm these results.
Acknowledgements
This research was supported in part by NIH R03 CA117509, NIH R01 CA48998 and NIH R25 CA94880.
References
- 1.Park SY, Murphy SP, Wilkens LR, et al. Calcium and vitamin D intake and risk of colorectal cancer: the Multiethnic Cohort Study. Am J Epidemiol. 2007;165:784–93. doi: 10.1093/aje/kwk069. [DOI] [PubMed] [Google Scholar]
- 2.Shin A, Li H, Shu XO, et al. Dietary intake of calcium, fiber and other micronutrients in relation to colorectal cancer risk: Results from the Shanghai Women’s Health Study. Int J Cancer. 2006;119:2938–42. doi: 10.1002/ijc.22196. [DOI] [PubMed] [Google Scholar]
- 3.Oh K, Willett WC, Wu K, Fuchs CS, Giovannucci EL. Calcium and vitamin D intakes in relation to risk of distal colorectal adenoma in women. Am J Epidemiol. 2007;165:1178–86. doi: 10.1093/aje/kwm026. [DOI] [PubMed] [Google Scholar]
- 4.Flood A, Peters U, Chatterjee N, et al. Calcium from diet and supplements is associated with reduced risk of colorectal cancer in a prospective cohort of women. Cancer Epidemiol Biomarkers Prev. 2005;14:126–32. [PubMed] [Google Scholar]
- 5.McCullough ML, Robertson AS, Rodriguez C, et al. Calcium, vitamin D, dairy products, and risk of colorectal cancer in the Cancer Prevention Study II Nutrition Cohort (United States) Cancer Causes Control. 2003;14:1–12. doi: 10.1023/a:1022591007673. [DOI] [PubMed] [Google Scholar]
- 6.Wu K, Willett WC, Fuchs CS, Colditz GA, Giovannucci EL. Calcium intake and risk of colon cancer in women and men. J Natl Cancer Inst. 2002;94:437–46. doi: 10.1093/jnci/94.6.437. [DOI] [PubMed] [Google Scholar]
- 7.Kampman E, Slattery ML, Caan B, Potter JD. Calcium, vitamin D, sunshine exposure, dairy products and colon cancer risk (United States) Cancer Causes Control. 2000;11:459–66. doi: 10.1023/a:1008914108739. [DOI] [PubMed] [Google Scholar]
- 8.Lipkin M. Preclinical and early human studies of calcium and colon cancer prevention. Ann N Y Acad Sci. 1999;889:120–7. doi: 10.1111/j.1749-6632.1999.tb08729.x. [DOI] [PubMed] [Google Scholar]
- 9.Newmark HL, Yang K, Lipkin M, et al. A Western-style diet induces benign and malignant neoplasms in the colon of normal C57Bl/6 mice. Carcinogenesis. 2001;22:1871–5. doi: 10.1093/carcin/22.11.1871. [DOI] [PubMed] [Google Scholar]
- 10.Vinas-Salas J, Biendicho-Palau P, Pinol-Felis C, Miguelsanz-Garcia S, Perez-Holanda S. Calcium inhibits colon carcinogenesis in an experimental model in the rat. Eur J Cancer. 1998;34:1941–5. doi: 10.1016/s0959-8049(98)00197-x. [DOI] [PubMed] [Google Scholar]
- 11.Buset M, Lipkin M, Winawer S, Swaroop S, Friedman E. Inhibition of human colonic epithelial cell proliferation in vivo and in vitro by calcium. Cancer Res. 1986;46:5426–30. [PubMed] [Google Scholar]
- 12.Pence BC, Buddingh F. Inhibition of dietary fat-promoted colon carcinogenesis in rats by supplemental calcium or vitamin D3. Carcinogenesis. 1988;9:187–90. doi: 10.1093/carcin/9.1.187. [DOI] [PubMed] [Google Scholar]
- 13.Chia V, Newcomb PA. Calcium and colorectal cancer: some questions remain. Nutr Rev. 2004;62:115–20. doi: 10.1111/j.1753-4887.2004.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 14.Norat T, Riboli E. Dairy products and colorectal cancer. A review of possible mechanisms and epidemiological evidence. Eur J Clin Nutr. 2003;57:1–17. doi: 10.1038/sj.ejcn.1601522. [DOI] [PubMed] [Google Scholar]
- 15.Cho E, Smith-Warner SA, Spiegelman D, et al. Dairy foods, calcium, and colorectal cancer: a pooled analysis of 10 cohort studies. J Natl Cancer Inst. 2004;96:1015–22. doi: 10.1093/jnci/djh185. [DOI] [PubMed] [Google Scholar]
- 16.Baron JA, Beach M, Mandel JS, et al. Calcium supplements for the prevention of colorectal adenomas. Calcium Polyp Prevention Study Group. N Engl J Med. 1999;340:101–7. doi: 10.1056/NEJM199901143400204. [DOI] [PubMed] [Google Scholar]
- 17.Bonithon-Kopp C, Kronborg O, Giacosa A, Rath U, Faivre J. Calcium and fibre supplementation in prevention of colorectal adenoma recurrence: a randomised intervention trial. European Cancer Prevention Organisation Study Group. Lancet. 2000;356:1300–6. doi: 10.1016/s0140-6736(00)02813-0. [DOI] [PubMed] [Google Scholar]
- 18.Hofstad B, Almendingen K, Vatn M, et al. Growth and recurrence of colorectal polyps: a double-blind 3-year intervention with calcium and antioxidants. Digestion. 1998;59:148–56. doi: 10.1159/000007480. [DOI] [PubMed] [Google Scholar]
- 19.Weingarten MA, Zalmanovici A, Yaphe J. Dietary calcium supplementation for preventing colorectal cancer and adenomatous polyps. Cochrane Database Syst Rev. 2008:CD003548. doi: 10.1002/14651858.CD003548.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grau MV, Baron JA, Sandler RS, et al. Prolonged effect of calcium supplementation on risk of colorectal adenomas in a randomized trial. J Natl Cancer Inst. 2007;99:129–36. doi: 10.1093/jnci/djk016. [DOI] [PubMed] [Google Scholar]
- 21.Wactawski-Wende J, Kotchen JM, Anderson GL, et al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. N Engl J Med. 2006;354:684–96. doi: 10.1056/NEJMoa055222. [DOI] [PubMed] [Google Scholar]
- 22.Twombly R. Negative Women’s Health Initiative findings stir consternation, debate among researchers. J Natl Cancer Inst. 2006;98:508–10. doi: 10.1093/jnci/djj172. [DOI] [PubMed] [Google Scholar]
- 23.Forman MR, Levin B. Calcium plus vitamin D3 supplementation and colorectal cancer in women. N Engl J Med. 2006;354:752–4. doi: 10.1056/NEJMe068006. [DOI] [PubMed] [Google Scholar]
- 24.Newmark HL, Wargovich MJ, Bruce WR. Colon cancer and dietary fat, phosphate, and calcium: a hypothesis. J Natl Cancer Inst. 1984;72:1323–5. [PubMed] [Google Scholar]
- 25.Canaff L, Hendy GN. Human calcium-sensing receptor gene. Vitamin D response elements in promoters P1 and P2 confer transcriptional responsiveness to 1,25-dihydroxyvitamin D. J Biol Chem. 2002;277:30337–50. doi: 10.1074/jbc.M201804200. [DOI] [PubMed] [Google Scholar]
- 26.Peters U, Chatterjee N, Yeager M, et al. Association of genetic variants in the calcium-sensing receptor with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2004;13:2181–6. [PubMed] [Google Scholar]
- 27.Speer G, Cseh K, Mucsi K, et al. Calcium-sensing receptor A986S polymorphism in human rectal cancer. Int J Colorectal Dis. 2002;17:20–4. doi: 10.1007/s003840100359. [DOI] [PubMed] [Google Scholar]
- 28.Fuszek P, Lakatos P, Tabak A, et al. Relationship between serum calcium and CA 19-9 levels in colorectal cancer. World J Gastroenterol. 2004;10:1890–2. doi: 10.3748/wjg.v10.i13.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.SeattleSNPs UW-FHCRC Variation Discovery Resource 2007, 2005.http://pga.gs.washington.edu
- 30.Slattery ML, Potter JD, Duncan DM, Berry TD. Dietary fats and colon cancer: assessment of risk associated with specific fatty acids. Int J Cancer. 1997;73:670–7. doi: 10.1002/(sici)1097-0215(19971127)73:5<670::aid-ijc10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 31.Clayton D. 2004 http://www-gene.cimr.cam.ac.uk/clayton/software/stata
- 32.Edwards S, Slattery ML, Mori M, et al. Objective system for interviewer performance evaluation for use in epidemiologic studies. Am J Epidemiol. 1994;140:1020–8. doi: 10.1093/oxfordjournals.aje.a117192. [DOI] [PubMed] [Google Scholar]
- 33.McDonald A, Van Horn L, Slattery M, et al. The CARDIA dietary history: development, implementation, and evaluation. J Am Diet Assoc. 1991;91:1104–12. [PubMed] [Google Scholar]
- 34.Slattery ML, Caan BJ, Duncan D, et al. A computerized diet history questionnaire for epidemiologic studies. J Am Diet Assoc. 1994;94:761–6. doi: 10.1016/0002-8223(94)91944-5. [DOI] [PubMed] [Google Scholar]
- 35.Liu K, Slattery M, Jacobs D, Jr., et al. A study of the reliability and comparative validity of the cardia dietary history. Ethn Dis. 1994;4:15–27. [PubMed] [Google Scholar]
- 36.NOAA/National Weather Service Climate Prediction Center: UV Index, Monthly Means and Maximums 2006, 2006.http://www.cpc.ncep.noaa.gov/products/stratosphere/uv_index/uv_meanmax.shtml
- 37.Sidney S, Jacobs DR, Jr., Haskell WL, et al. Comparison of two methods of assessing physical activity in the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Am J Epidemiol. 1991;133:1231–45. doi: 10.1093/oxfordjournals.aje.a115835. [DOI] [PubMed] [Google Scholar]
- 38.Slattery ML, Jacobs DR., Jr. Assessment of ability to recall physical activity of several years ago. Ann Epidemiol. 1995;5:292–6. doi: 10.1016/1047-2797(94)00095-b. [DOI] [PubMed] [Google Scholar]
- 39.Willett W, Stampfer MJ. Total energy intake: implications for epidemiologic analyses. Am J Epidemiol. 1986;124:17–27. doi: 10.1093/oxfordjournals.aje.a114366. [DOI] [PubMed] [Google Scholar]
- 40.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 41.University of California Santa Cruz Genome Browser 2006, 2006.http://genome.ucsc.edu
- 42.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunyaev S, Ramensky V, Koch I, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 44.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 45.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–71. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–13. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 47.Pesole G, Liuni S. Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Trends Genet. 1999;15:378. doi: 10.1016/s0168-9525(99)01795-3. [DOI] [PubMed] [Google Scholar]
- 48.Chakrabarty S, Radjendirane V, Appelman H, Varani J. Extracellular calcium and calcium sensing receptor function in human colon carcinomas: promotion of E-cadherin expression and suppression of beta-catenin/TCF activation. Cancer Res. 2003;63:67–71. [PubMed] [Google Scholar]
- 49.Kallay E, Bajna E, Wrba F, et al. Dietary calcium and growth modulation of human colon cancer cells: role of the extracellular calcium-sensing receptor. Cancer Detect Prev. 2000;24:127–36. [PubMed] [Google Scholar]
- 50.Kallay E, Kifor O, Chattopadhyay N, et al. Calcium-dependent c-myc proto-oncogene expression and proliferation of Caco-2 cells: a role for a luminal extracellular calcium-sensing receptor. Biochem Biophys Res Commun. 1997;232:80–3. doi: 10.1006/bbrc.1997.6225. [DOI] [PubMed] [Google Scholar]
- 51.Lamprecht SA, Lipkin M. Chemoprevention of colon cancer by calcium, vitamin D and folate: molecular mechanisms. Nat Rev Cancer. 2003;3:601–14. doi: 10.1038/nrc1144. [DOI] [PubMed] [Google Scholar]
- 52.Buras RR, Shabahang M, Davoodi F, et al. The effect of extracellular calcium on colonocytes: evidence for differential responsiveness based upon degree of cell differentiation. Cell Prolif. 1995;28:245–62. doi: 10.1111/j.1365-2184.1995.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 53.Brown EM, MacLeod RJ. Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev. 2001;81:239–97. doi: 10.1152/physrev.2001.81.1.239. [DOI] [PubMed] [Google Scholar]
- 54.Gama L, Baxendale-Cox LM, Breitwieser GE. Ca2+-sensing receptors in intestinal epithelium. Am J Physiol. 1997;273:C1168–75. doi: 10.1152/ajpcell.1997.273.4.C1168. [DOI] [PubMed] [Google Scholar]
- 55.Mitsuma T, Rhue N, Kayama M, et al. Distribution of calcium sensing receptor in rats: an immunohistochemical study. Endocr Regul. 1999;33:55–9. [PubMed] [Google Scholar]
- 56.Cheng SX, Okuda M, Hall AE, Geibel JP, Hebert SC. Expression of calcium-sensing receptor in rat colonic epithelium: evidence for modulation of fluid secretion. Am J Physiol Gastrointest Liver Physiol. 2002;283:G240–50. doi: 10.1152/ajpgi.00500.2001. [DOI] [PubMed] [Google Scholar]
- 57.Buchan AM, Squires PE, Ring M, Meloche RM. Mechanism of action of the calcium-sensing receptor in human antral gastrin cells. Gastroenterology. 2001;120:1128–39. doi: 10.1053/gast.2001.23246. [DOI] [PubMed] [Google Scholar]
- 58.Chattopadhyay N, Cheng I, Rogers K, et al. Identification and localization of extracellular Ca(2+)-sensing receptor in rat intestine. Am J Physiol. 1998;274:G122–30. doi: 10.1152/ajpgi.1998.274.1.G122. [DOI] [PubMed] [Google Scholar]
- 59.Hobson SA, Wright J, Lee F, et al. Activation of the MAP kinase cascade by exogenous calcium-sensing receptor. Mol Cell Endocrinol. 2003;200:189–98. doi: 10.1016/s0303-7207(01)00749-3. [DOI] [PubMed] [Google Scholar]
- 60.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–22. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–8. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 62.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–88. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 63.McMichael AJ, Potter JD. Host factors in carcinogenesis: certain bile-acid metabolic profiles that selectively increase the risk of proximal colon cancer. J Natl Cancer Inst. 1985;75:185–91. [PubMed] [Google Scholar]
- 64.Brown EM. The extracellular Ca2+-sensing receptor: central mediator of systemic calcium homeostasis. Annu Rev Nutr. 2000;20:507–33. doi: 10.1146/annurev.nutr.20.1.507. [DOI] [PubMed] [Google Scholar]
- 65.Bronner F, Pansu D. Nutritional aspects of calcium absorption. J Nutr. 1999;129:9–12. doi: 10.1093/jn/129.1.9. [DOI] [PubMed] [Google Scholar]
- 66.Karbach U, Feldmeier H. The cecum is the site with the highest calcium absorption in rat intestine. Dig Dis Sci. 1993;38:1815–24. doi: 10.1007/BF01296104. [DOI] [PubMed] [Google Scholar]
- 67.Hartman TJ, Albert PS, Snyder K, et al. The association of calcium and vitamin D with risk of colorectal adenomas. J Nutr. 2005;135:252–9. doi: 10.1093/jn/135.2.252. [DOI] [PubMed] [Google Scholar]
- 68.Terry P, Baron JA, Bergkvist L, Holmberg L, Wolk A. Dietary calcium and vitamin D intake and risk of colorectal cancer: a prospective cohort study in women. Nutr Cancer. 2002;43:39–46. doi: 10.1207/S15327914NC431_4. [DOI] [PubMed] [Google Scholar]
- 69.Marcus PM, Newcomb PA. The association of calcium and vitamin D, and colon and rectal cancer in Wisconsin women. Int J Epidemiol. 1998;27:788–93. doi: 10.1093/ije/27.5.788. [DOI] [PubMed] [Google Scholar]
- 70.Kearney J, Giovannucci E, Rimm EB, et al. Calcium, vitamin D, and dairy foods and the occurrence of colon cancer in men. Am J Epidemiol. 1996;143:907–17. doi: 10.1093/oxfordjournals.aje.a008834. [DOI] [PubMed] [Google Scholar]
- 71.Peters RK, Pike MC, Garabrant D, Mack TM. Diet and colon cancer in Los Angeles County, California. Cancer Causes Control. 1992;3:457–73. doi: 10.1007/BF00051359. [DOI] [PubMed] [Google Scholar]
- 72.Stemmermann GN, Nomura A, Chyou PH. The influence of dairy and nondairy calcium on subsite large-bowel cancer risk. Dis Colon Rectum. 1990;33:190–4. doi: 10.1007/BF02134177. [DOI] [PubMed] [Google Scholar]
- 73.Lin J, Zhang SM, Cook NR, et al. Intakes of calcium and vitamin D and risk of colorectal cancer in women. Am J Epidemiol. 2005;161:755–64. doi: 10.1093/aje/kwi101. [DOI] [PubMed] [Google Scholar]
- 74.Wright J, Wang C, Kennedy-Stephenson J, Ervin R. Dietary intake of ten key nutrients for public health, United States: 1999–2000. National Center for Health Statistics; Hyattsville, Maryland: 2003. [PubMed] [Google Scholar]