Abstract
Objective
The present study evaluated differences in gene expression associated with age and gender in the human parotid gland.
Design
Parotid gland tissue was analyzed using the Affymetrix® GeneChip® HGU133plus2.0 array.
Results
Differential gene expression, defined as a statistically significant difference with a 1.5 fold or greater change, was detected in 787 gene probe sets; 467 (~59%) showed higher expression in females. Several genes associated with saliva secretion were differentially expressed in male and female parotid glands including vesicle-associated membrane protein 3 VAMP3, synaptosomal-associated protein SNAP23, RAS oncogene family member RAB1A and the syntaxin binding protein STXBP1. Evaluation of gene expression in the youngest and the oldest female subjects revealed that the expression of 228 probe sets were altered during aging; 155 genes were up-regulated in the aged female parotid gland. However, of the genes that were altered during aging, 22 of the 30 probes (73%) classified as being associated with immune responses were down-regulated in the aged parotid gland. A panel of differentially expressed, age- and gender-related genes was selected for validation by quantitative, real-time RT-PCR. Comparable differences in gene expression were detected by both Affymetrix array and quantitative, real-time RT-PCR methods.
Conclusions
Our data suggest that salivary gland function may be adversely affected in the aged population due, at least in part, to the altered regulation of several categories of genes. Moreover, the gender specific differences in gene expression identified in the present study correlate with the previously observed sexual dimorphism in salivary gland function.
Keywords: Human parotid, Salivary gland, Gene expression
INTRODUCTION
Previous studies suggest that there may be age and gender related differences in salivary gland function (1–5). However, the limited and often conflicting information available from healthy populations makes it difficult to confirm these differences (6–9). Saliva is produced and secreted into the oral cavity by the exocrine salivary glands. Humans possess three major pairs of salivary glands (parotid, submandibular and sublingual) and several types of minor salivary glands scattered throughout the oral cavity. The majority of saliva (>80%) is generated by the two largest of these glands, the parotid and submandibular glands (10, 11). Recent studies show that saliva contains well over one thousand different unique proteins, the functions for the majority of which are yet to be determined (12).
Salivation is a highly regulated process which occurs at a relatively slow rate between meals, with almost no secretion during sleep (13). The importance of saliva to oral health is most evident in subjects suffering from severe salivary gland hypofunction commonly associated with the autoimmune disease Sjögren’s syndrome, radiation therapy of head and neck cancers, and numerous types of medications. In these cases, there is a dramatic increase in both oral and systemic disease (14). Thus, without adequate saliva output, oral and pharyngeal health declines along with quality of life.
Persistent dry mouth is also a common symptom in aged individuals, although the mechanisms involved are not well understood. Dry mouth in the elderly is frequently associated with the increased use of medications and the functional disturbances associated with these medications. Both age and gender associated differences in the structure and function of salivary glands have been identified (1, 4). Examples of such differences include a decrease in gland size and weight (15), decreased saliva flow rate (1–3), and an increased concentration of immunoglobulin A (IgA) (3). In humans, decreases in protein synthesis (16) and salivary flow rate (1, 2, 4, 5) have been reported. Given these age related differences in salivary gland structure and function, it is expected that significant changes in gene expression must occur. However, other studies have failed to demonstrate a relationship between age and decreased function (6–9). Thus, the limited and often conflicting information available from healthy populations makes it difficult to confirm these differences. The purpose of the present study was to evaluate and compare differences in gene expression associated with age and gender in the human parotid gland.
MATERIALS and METHODS
Human Parotid Gland Tissue
Human parotid glands were obtained from 32 otherwise healthy male (n=13) and female (n=19) subjects (19–85 years of age) scheduled to have parotid surgery because their gland contained a benign tumor that required removal of all or a large portion of the gland. All samples were pathologically confirmed to be benign salivary gland tumors (pleomorphic adenoma, mixed tumor or Warthin’s tumor). Subjects were excluded who indicated that they experienced dry mouth or took medications known to adversely affect salivary gland function. Much of the normal tissue surrounding the tumor is not used for diagnostic evaluation of the sample. This discarded tissue was collected immediately after surgical excision and transported in ice-cold physiological saline to the laboratory where the tissue was frozen in liquid N2. Tissue was obtained and used as approved by the University of Rochester Institutional Review Board, or in the case of 3 samples, obtained through the Cooperative Human Tissue Network (CHTN). The functional properties of the parotid tissue obtained by these criteria appear normal in all respects (17, 18).
RNA Isolation and Array Analysis
Total RNA from parotid gland tissues was treated with RNase free DNase (Qiagen) and isolated by affinity chromatography according to the manufacturer’s protocol (RNeasy kit (Qiagen, Valencia, CA). RNA (3 µg) was subjected to 1 round of linear amplification with the RiboAmp™ RNA Amplification kit (ARCTURUS, Mountain View CA) and biotinylated using GeneChip® Expression 3’-Amplification reagents for IVT labeling (Affymetrix). Before hybridization, 13 µg of labeled RNA was fragmented using 5X fragmentation Buffer (Affymetrix). RNA quality was monitored before and after amplification, as well after fragmentation (2100 bioanalyzer, Agilent Technologies). Hybridization to the Human Genome U133 Plus 2.0 Array as well as imagine scanning (Affymetrix, Santa Clara, CA, USA) was performed by the Microarray Core Facility at the University of California, Los Angeles, according to standard protocols provided by Affymetrix.
Array Data and Statistic Analysis
The fluorescence intensities of the arrays were measured by Array Suite 5.0 software (Affymetrix). The data were imported into DNA-Chip Analyzer software (Affymetrix) for normalization and model-based analysis (19). A detection p-value was obtained for each probe set, and any probe sets with p < 0.04 were assigned as a "present" call, indicating that the matching gene transcript was reliably detected (Statistical algorithms description document. Affymetrix, 2002). The raw data were then exported to Microsoft Excel software for data sorting and mining.
The GeneSifter® array data analysis system (VizX Labs LLC, Seattle, WA) was used to identify age and gender related differences in gene expression of the human parotid gland. The Affymetrix non-normalized data (CEL files) were transferred into GeneSifter. Expression measurements were derived using RMA (robust multiple average) and filtering criteria of a 1.5 or greater fold change (20). Statistical significance was determined by Student’s t-test, and data were corrected for multiple testing using the method of Benjamini and Hochberg (21). Only the probe sets which passed the quality filtering with p values <0.05 were included in the analysis.
Pathway Analysis
Genes with significantly different expression were overlaid using GeneSifter software onto Ontological pathways (http://www.geneontology.org/) (22) and KEGG pathways (http://www.genome.jp/kegg/) (23). The ontological and KEGG pathway analyses provided detailed data on individual genes in the context of that gene's role in described biological processes, molecular functions, and cellular components. Pathways were considered significantly altered from the control gene expression profiles if the z-score for that pathway was less than −2 or greater than 2. z-Scores were calculated in GeneSifter using the following formula:
where R = total number of genes meeting selection criteria, N = total number of genes measured, r = number of genes meeting selection criteria with the specified GO term, and n = total number of genes measured with the specific GO term (24). The data from the individual arrays (n = 13) are accessible for download through the National Center for Biotechnology Information’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) through series accession number (GSE8764).
RT-PCR and Q-PCR
Quantitative PCR (Q-PCR) was used to validate microarray results. First-strand complementary DNA (cDNA) was synthesized from 1 µg of total RNA in a 20 µl reaction using the iScript™ cDNA synthesis kit (Bio-Rad, Hercules, CA). A primer set for each gene was generated using GeneFisher software (http://bibiserv.techfak.uni-bielefeld.de/genefisher/). The primers were synthesized commercially (Integrated DNA Technologies, Coralville, IA). Q-PCR was performed in triplicate using a 96-well iCycler IQ™ Real Time PCR Detection System (Bio-Rad, Hercules, CA) in 25 µl total volume containing 12.5 µl of 2XSYBR Green Master Mix , 0.5 to 1.0 µl of cDNA and 0.5 µl and 2 ng/µl of gene specific primers (Table 1). Q-PCR amplification was carried out by 40 cycles of 30 sec at 95°C and 30 sec at 60°C. Dissociation curves were monitored (60°C for 10 sec to 95°C in 0.5°C/10 sec increments) to ensure the absence of secondary PCR product. The predicted sizes of the PCR products were verified by agarose gel electrophoresis. In most cases, the PCR products were also sequence verified. The endogenous control, L32, was measured simultaneously for each sample. The PCR efficiency of the reaction was measured with L32 primers using serial dilution of cDNA (1:1, 1:2, 1:4, 1:8 and 1:16). Q-PCR amplification curves were analyzed with iCycler IQ Software version 3.1.7050. For relative quantification of gene expression, the comparative threshold cycle (Ct) method was used (described in User Bulletin 2 for ABI PRISM® 7700 Sequence Detection Systems). The value obtained from Ct represents the PCR cycle at which an increase in fluorescence signal can be detected above background for the particular gene. The Ct values of endogenous control (L32) were subtracted from that of each gene of interest Ct values to derive the ΔCt value. The relative expression of the gene of interest, ΔΔCt, was then evaluated by subtracting the ΔCt of control sample from the compared sample, e.g., male (control) to female (compared) or young (control) to old (compared). The fold difference was calculated as 2-ΔΔCt.
Table 1.
Oligonucleotide primers used for Q-PCR
| Accession No. | Gene ID | Forward (5'–3') | Reverse (5'–3') |
|---|---|---|---|
| NM_000738 | CHRM1 | TTCCTGGGAGTGGGAGTCAAG | ATTGGGGAGCTCACAGGAGAG |
| NM_138455 | CTHRC1 | TCGCACTTCTTCTGTGGAAGG | TGCGAGAAACTGAATTCCATCC |
| AF332225 | CYORF15B | GGCAGTTTCTTAGGCTGTGAC | TTGTTTCCAATGCTAGCCAGAG |
| NM_004660 | DDX3Y | ACTGATAGGAAGGTCCACATCC | AATACTGCTGGCTGGTAAAACC |
| NM_152679 | SLC10A4 | TGTGGAGATACACAGGAGCTTC | GGCTTCACGTTAGCCATTCC |
| NM_139266 | STAT1 | CAAACCTCAAGCCAGCCTTG | GGCAGTAACACGGGGATCTC |
| NR_001564 | XIST | AAACAAGGTGTTGTGGTCTTCC | TCAGCTGTCAGTGATCTAATGC |
| BC033974 | ZFY | CTTCCCTCTCACTCCTGGTAC | CAGGCAGAAGAAAGAATCAGCA |
| NM_001565 | CXCL10 | GAGGTGCTATGTTCTTAGTGGATG | CTGAAAGAATTTGGGCCCCTTG |
| NM_002122 | HLA-DQA1 | GCTATATCCCCTCAGAGCTCAC | AGTCAGCCCTGGATGAAAGATG |
RESULTS
Gene Expression in Human Parotid Glands Measured by Array
RNA was isolated from the parotid glands of 8 female and 5 male subjects 19 to 71 years of age and analyzed by microarray. The Affymetrix® GeneChip® HGU133 Plus 2.0 array contains about 54,000 probe sets representing over 18,400 transcripts and variants, including 14,500 well-characterized human genes. The percent array outliers (19) were in the range of 0.12 to 0.31, and the percent of a single outlier was in the range of 0.11 and 1.02 (19). The percent of “present” call varied from array to array, ranging from 41.2% to 52.5%. About 21% of the probe sets were not detected on any of the 13 arrays. There was a common present call for about 26% of the probe sets on all 13 arrays. An additional 16% of the positive probe sets were detected on at least ten of the 13 arrays. Thus, nearly 37% of the probes sets were “present” on 10 of the 13 samples analyzed, which is similar to the number of genes expressed in other organ systems (25, 26).
Age Related Differences in Human Female Parotid Gland Gene Expression
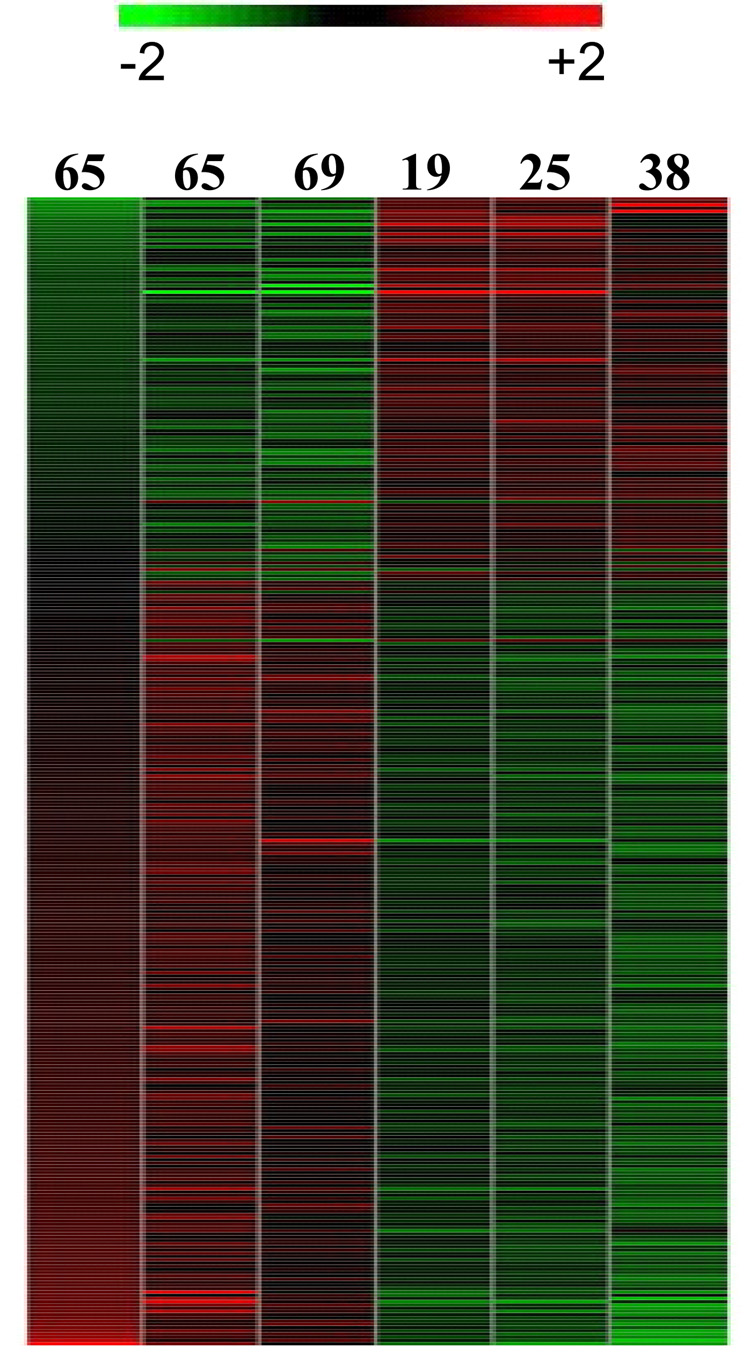
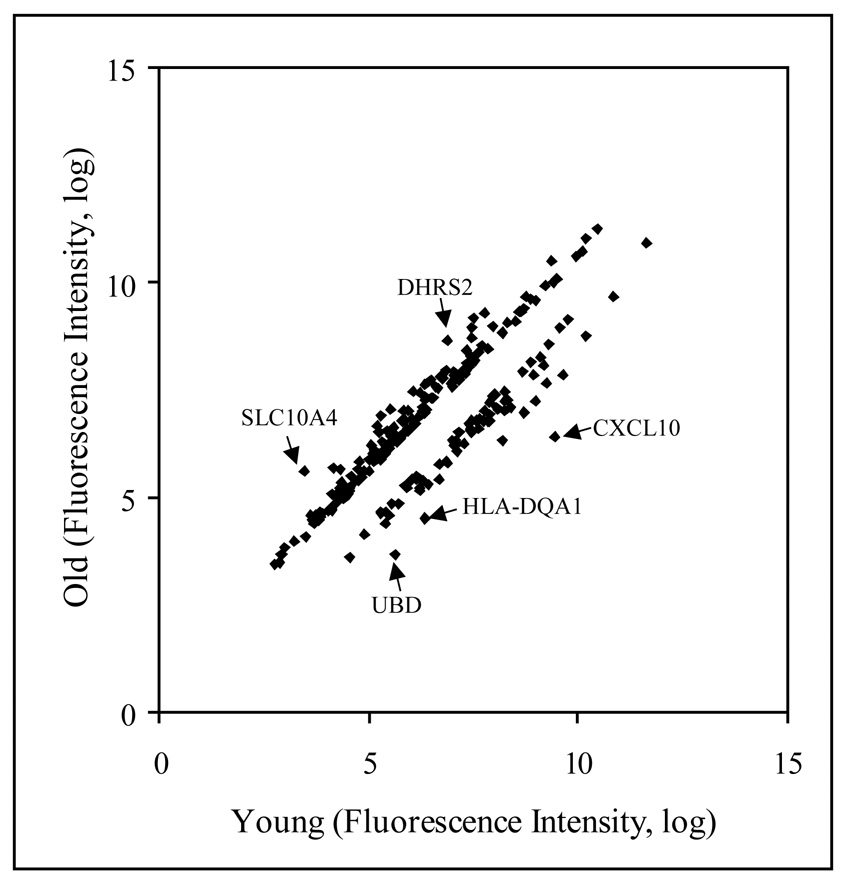
Array data from human female parotid glands were compared for differences in gene expression. Data were analyzed by GeneSifter array data analysis system using RMA (robust multi-array analysis) and filtering criteria of a 1.5 or greater fold change (see METHODS). To eliminate gender differences in gene expression, only female samples were used for this analysis. Samples from 3 young females (19, 25 and 38 years old) and 3 older females (65, 65 and 69 years old) were compared (Figure 1). The expression of 228 probe sets showed differential expression between these two age groups; the signal on 155 (68%) of these probe sets increased in aged parotid glands (Figure 2, Supplement Table A). Probe sets representing 10 unique genes showed a 3 fold or more difference in expression during aging including e.g., CXCL10, UBD, HLA-DQA1 (Table 2, also see full Supplement Table 1). A few examples of highly differentially expressed genes are indicated by arrows in Figure 2. The differentially expressed genes are involved in numerous biological functions such as chemokine (CXCL10), electron transport (DHRS2), ion transport (SLC10A4), antigen processing (HLA-DQA1), proteolysis (UBD).
Figure 1.
Heat map of 228 differentially expressed genes comparing RNA samples from female human parotid of different age groups. The age is noted on each lane. Red represents relative expression greater than the median expression level across all samples, and green represents an expression level lower than the median. Black indicates intermediate expression.
Figure 2.
Scattered plot analysis of 228 genes, which have at least 1.5 fold difference in their expression in between young (19, 25 and 38 years of age) and old (65, 65 and 69 years of age) female parotid glands.
Table 2.
Gene expression ratio of older (65, 65 and 69 years) to younger (19, 25 and 38 years) (O/Y) female parotid glands*
| Gene Name | Acc. No. | Gene ID | Ratio (O/Y) | Direction | Ontology |
|---|---|---|---|---|---|
| Cytokine subfamily B (Cys-X-Cys), member 10 | NM_001565 | CXCL10 | 8.37 | Down | Chemokine |
| Solute carrier family 10 member 4 | AI421796 | SLC10A4 | 4.41 | Up | Ion transport |
| Diubiquitin D | NM_006398 | UBD | 3.89 | Down | Proteolysis |
| Histocompatibility complex, class II, DQ alpha 1 | BG397856 | HLA-DQA1 | 3.5 | Down | Antigen processing |
| Interferon, alpha-inducible protein | NM_022873 | IFI6 | 3.49 | Down | Release of cytochrome c from mitochondria |
| Short-chain alcohol dehydrogenase family member | NM_005794 | DHRS2 | 3.38 | Up | Electron transport |
| Interferon-stimulated transcription factor 3, gamma | NM_006084 | ISGF3G | 3.34 | Down | Regulation of transcription |
| Signal transducer and activator of transcription 1, 91kDa | BC002704 | STAT1 | 3.31 | Down | Regulation of transcription |
| Interferon-stimulated protein, 15 kDa | NM_005101 | ISG15 | 3.05 | Down | Response to other organism |
| Ependymin related protein 1 | BC000686 | EPDR1 | 3 | Up | Ion binding |
| Cysteine/tyrosine-rich 1 | H06649 | CYYR1 | 2.92 | Up | |
| Periostin, osteoblast specific factor | D13665 | POSTN | 2.86 | Up | Skeletal development |
| Collagen triple helix repeat containing 1 | AA584310 | CTHRC1 | 2.82 | Up | Phosphate transport |
| Histone deacetylase 9 | BM726008 | HDAC9 | 2.73 | Up | Negative regulation of transcription |
| Phosphodiesterase 5A, cGMP-specific | BF221547 | PDE5A | 2.68 | Up | Signal transduction |
| Immunoglobulin heavy constant gamma 1 | BC001872 | IGHG1 | 2.66 | Down | Antigen processing |
| Stearoyl-CoA desaturase | AA678241 | SCD | 2.49 | Down | Lipid metabolic process |
| Rho GTPase activating protein 6 | NM_001174 | ARHGAP6 | 2.4 | Up | Regulation of catalytic activity |
| Glycoprotein (transmembrane) nmb (GPNMB) | NM_002510 | GPNMB | 2.37 | Up | Negative regulation of cellular process |
| Interferon regulatory factor 7 | NM_004030 | IRF7 | 2.36 | Down | Negative regulation of transcription |
| Four and a half LIM domains 1 | AF063002 | FHL1 | 2.31 | Up | Metal Binding |
| Lysosomal associated protein transmembrane 4 beta | AW149681 | LAPTM4B | 2.31 | Up | Transport |
| Cytochrome P450, family 4, subfamily B, polypeptide 1 | J02871 | CYP4B1 | 2.26 | Up | Electron transport |
| Frizzled-related protein | U91903 | FRZB | 2.23 | Up | Cell communication |
| Four and a half LIM domains 1 (FHL1) | NM_001449 | FHL1 | 2.18 | Up | Metal Binding |
| Solute carrier family 1 , member 1 | AW235061 | SLC1A1 | 2.16 | Up | Transport |
| Epithelial stromal interaction 1 | AA781795 | EPSTI1 | 2.15 | Down | |
| Guanylate binding protein 1, interferon-inducible | AW014593 | GBP1 | 2.15 | Down | Immune response |
| Tubulin tyrosine ligase-like family, member 7 | NM_024686 | TTLL7 | 2.14 | Up | Protein modification |
| NGFRAP1-like 1 | AV726956 | NGFRAP1L1 | 2.13 | Down | |
| Fatty acid synthase | AI954041 | FASN | 2.12 | Down | Fatty acid biosynthetic process |
| Guanylate binding protein 1, interferon-inducible | BC002666 | GBP1 | 2.1 | Down | Immune response |
| Interferon induced transmembrane protein 3 (1-8U) | BF338947 | IFITM3 | 2.1 | Down | Immune response |
| Asparaginase like 1 | NM_025080 | ASRGL1 | 2.09 | Up | Glycoprotein catabolic process |
| Myxovirus (influenza virus) resistance 2 | NM_002463 | MX2 | 2.09 | Down | Defense response |
| Solute carrier organic anion transporter, member 1A2 | NM_021094 | SLCO1A2 | 2.09 | Up | Transport |
| Interferon induced transmembrane protein 1 | AA749101 | IFITM1 | 2.08 | Down | Immune response |
| Plasminogen activator, tissue | NM_000930 | PLAT | 2.07 | Up | Protein modification |
| Solute carrier family 25, member 34 | AU151211 | SLC25A34 | 2.07 | Up | Transport |
| ATP-binding cassette, member 2 (ABCB2) | NM_000593 | TAP1 | 2.03 | Down | Response to stimulus |
| Low density lipoprotein receptor | NM_000527 | LDLR | 2.03 | Down | Protein Biosysnthesis |
| Cysteine/tyrosine-rich 1 | AI458003 | CYYR1 | 2.02 | Up | |
| NLR family, CARD domain containing 5 | AA005023 | NLRC5 | 2.02 | Down | Defense response |
| Lysosomal associated protein transmembrane 4 beta | NM_018407 | LAPTM4B | 2.01 | Up | Transport |
| Monocyte to macrophage differentiation-associated | NM_012329 | MMD | 2.01 | Up | Cytolysis |
| Peroxisomal biogenesis factor 6 | NM_000287 | PEX6 | 2.01 | Down | Peroxisome organization |
| Integrin beta 1 binding protein 1 | NM_004763 | ITGB1BP1 | 2 | Up | Cell adhesion |
Genes listed had signal intensity of >5.0 in at least one group, expression ratio of >2.0 (between glands), p value <0.05, with known gene identity.
Interestingly, a large number of the differentially expressed genes include those known to be involved in immune responses (Table 3). The very high z-score (11.72, Table 3) indicates that the older population may have an altered immune system. Out of the 522 probe sets on the HGU133A Plus 2.0 array known to be associated with the immune system, 30 probe sets were differentially expressed. The expression decreased on 22 probe sets while expression increased on 8 probe sets in the older population. A list of the 30 differentially expressed probe sets involved in immune response is given in Table 4. The expression of both HLA-DQA1 and HLA-DQB1 are decreased in the parotid gland of the aged female, as well as Chemokine (C-X-C motif) ligand 10 (CXCL10). Several other proteins (e.g., IRF6, IRF7, GBP1, IFITM1, IFITM2, PSMB8 and PSMB9), which are known to be involved in different immune response pathways, showed altered expression in the aged population (Table 4).
Table 3.
Differential gene expression in older (69,65 and 65 years) compared to younger (19, 25 and 38) female parotid glands and their ontological categorization based on their immunity response
| Ontology | Diff exp genesa | Up-regb | Down-regb | Tot on arrayc | z-Score upd | z-Score downd |
|---|---|---|---|---|---|---|
| Immune response | 28 | 4 | 24 | 522 | −1.14 | 11.72 |
| Defense response | 16 | 6 | 10 | 476 | −0.13 | 4.14 |
| Antigen processing and presentation | 9 | 0 | 9 | 52 | −0.84 | 15.24 |
| Response to biotic stimulus | 9 | 0 | 9 | 215 | −1.37 | 8.58 |
| Response to other organism | 7 | 1 | 6 | 151 | −0.72 | 5.23 |
| Response to virus | 6 | 0 | 6 | 84 | −1.07 | 7.57 |
Diff exp genes indicates the total number of genes differentially expressed on the array in that category.
Up-reg and down-reg indicate the total number of up and down regulated genes respectively in older population.
Tot on array indicates the total number of genes on the array in that onctological category.
z-score-up and z-score-down indictae the z-score for that category.
Table 4.
Differentially expressed genes known to be involved in the immune response in older (65, 65 and 69 years) compared to younger (19, 25 and 38 years) female parotid glands
| Gene Name | Accession No. | Gene ID | Ratio | Direction |
|---|---|---|---|---|
| Chemokine (C-X-C motif) ligand 10 | NM_001565 | CXCL10 | 8.37 | Down |
| Ubiquitin D | NM_006398 | UBD | 3.89 | Down |
| Major histocompatibility complex, class II, DQ alpha 1 | BG397856 | HLA-DQA1 | 3.5 | Down |
| Interferon, alpha-inducible protein 6 | NM_022873 | IFI6 | 3.49 | Down |
| Interferon-stimulated transcription factor 3, gamma 48kDa | NM_006084 | ISGF3G | 3.34 | Down |
| interferon-stimulated protein, 15 kDa | NM_005101 | ISG15 | 3.05 | Down |
| Histone deacetylase 9 | BM726008 | HDAC9 | 2.73 | Up |
| interferon regulatory factor 7 | NM_004030 | IRF-7 | 2.36 | Down |
| guanylate binding protein 1, interferon-inducible, 67kD | AW014593 | GBP1 | 2.15 | Down |
| Guanylate binding protein 1, interferon-inducible | BC002666 | GBP1 | 2.1 | Down |
| Interferon induced transmembrane protein 3 | BF338947 | IFITM3 | 2.1 | Down |
| Myxovirus (influenza virus) resistance 2 | NM_002463 | MX2 | 2.09 | Down |
| Interferon induced transmembrane protein 1 | AA749101 | IFITM1 | 2.08 | Down |
| Transporter 1, ATP-binding cassette, sub-family B | NM_000593 | TAP1 | 2.03 | Down |
| Major histocompatibility complex, class II, DQ beta 1 | AI583173 | HLA-DQB1 | 1.99 | Down |
| Myeloid leukemia factor 1 | NM_022443 | MLF1 | 1.97 | Up |
| Proteasome subunit, beta type, 9 | NM_002800 | PSMB9 | 1.97 | Down |
| NCK adaptor protein 1 | NM_006153 | NCK1 | 1.95 | Up |
| Suppressor of cytokine signaling 5 | AW664421 | SOCS5 | 1.95 | Up |
| Interferon regulatory factor 1 | NM_002198 | IRF-1 | 1.89 | Down |
| Secreted and transmembrane 1 | BF939675 | SECTM1 | 1.82 | Down |
| Leptin | NM_000230 | LEP | 1.74 | Up |
| Clusterin | M25915 | CLU | 1.67 | Up |
| CD74 molecule | K01144 | CD74 | 1.63 | Down |
| Mucosa associated lymphoid tissue lymphoma translocation ge | NM_006785 | MALT1 | 1.63 | Up |
| Proteasome subunit, beta type, 9 | AI375915 | PSMB9 | 1.62 | Down |
| Proteasome subunit, beta type, 8 | U17496 | PSMB8 | 1.61 | Down |
| Tumor necrosis factor (ligand) superfamily, member 13b | AF134715 | TNFSF13B | 1.58 | Down |
| Interferon induced transmembrane protein 2 | NM_006435 | IFITM2 | 1.55 | Down |
| Chemokine (C-C motif) receptor 2 | NM_000647 | CCR2 | 1.5 | Up |
The older population also showed altered expression of several ion transporters and neurotransmitter receptors known to be involved in saliva secretion, e.g., the cholinergic muscarinic type 1 receptor CHRM1 and the K channel KCNJ2 showed lower expression in the aged (Supplement Table A). Other ion transporters and channels, such as, SLC10A4, CTHRC1 (Phosphate/organic transporter), SLC21A3, SLC01A2, SLC24A3 SLC30A9, SCL39A10 and CLCN3 also showed differential expression (Supplement Table A) [the data from the 13 individual arrays are accessible for download through the National Center for Biotechnology Information’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) through series accession number (GSE8764)].
Gender Related Differences in Human Parotid Tissue
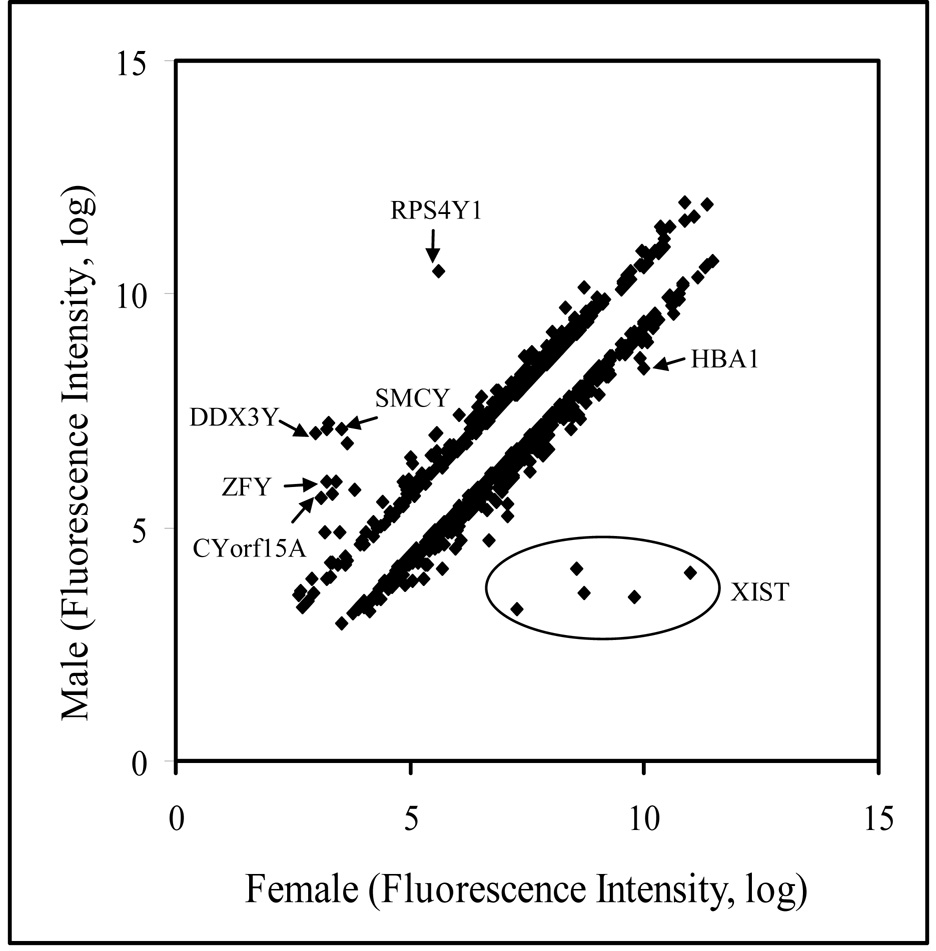
To determine whether gender-related gene expression differences exist in the human parotid gland, eight female samples (19, 25, 38, 41, 49, 65, 65 and 69 years of age) and five male samples (42, 59, 62, 70 and 71 years old) were analyzed by microarray. Examination of the array data by GeneSifter software demonstrated that gender has a very significant influence on gene expression in the parotid gland with 787 probe sets showing differential expression. Table 5 presents the gene expression differences between male and female parotid samples. Male tissues showed higher expression with 320 probe sets, while, 467 probe sets were preferentially expressed at a higher level in females. Ten unique gene probe sets were expressed at greater than a 10-fold difference with an additional 79 gene probes were over-expressed at more than a two-fold difference. Not surprisingly, the greatest differences in expression levels (up to 124-fold) were observed in the genes linked to the X and Y chromosomes (Figure 3 and Table 6). Twenty-five probe sets representing genes on the X chromosome were differentially expressed. Of these, 19 were found at higher levels in female glands, while 6 probe sets were slightly higher in male tissue. HBA2 is involved in oxygen transport while XIST regulates X chromosome inactivation (Figure 3). Multiple probe sets for the XIST gene are present on the array which all show much higher expression in females (Figure 3). All 17 of the probe sets for genes on the Y chromosome showed higher expression in male samples and almost no signal was detected in female samples. Representative examples are shown by arrows in Figure 3. These Y chromosome-specific genes are involved in various biological functions such as protein biosynthesis (RPS4Y1), nucleotide binding (DDX3Y), transcription regulation (SMCY, ZFY).
Table 5.
Differential expression of probe sets in male (n=5) and female (n=8) parotid glands*
| Expression Threshhold | Differentially Expressed Genes | Genes Up-reguatted in Male | Genes Up-regulated in Female |
|---|---|---|---|
| >1.5 fold | 787 | 320 | 467 |
| >1.5 fold and <2 | 698 | 280 | 418 |
| >2 fold and <10 | 79 | 35 | 44 |
| >10 fold | 10 | 5 | 5 |
A student t-test was applied, with the p value <0.05.
Figure 3.
Scattered plot analysis of 787 genes, which have at least 1.5 fold differences in their expression in between male and female parotid glands.
Table 6.
Gender related differential gene expression in human parotid gland*
| Higher Expression in Female | ||||
|---|---|---|---|---|
| Gene Name | Accession No. | Gene ID | Ratio | Ontology |
| X (inactive)-specific transcript | NR_001564 | XIST | 124.03 | |
| Zinc finger, CCHC domain containing 2 | NM_017742 | ZCCHC2 | 3.52 | Metal Ion Binding |
| Pinin, desmosome associated protein | NM_002687 | PNN | 3.03 | Transcription regulation |
| Hemoglobin, alpha1 | NM_000517 | HBA1 | 3.01 | Oxygen transport |
| PRO1073 protein | NM_014086 | PRO1073 | 2.62 | Unknown |
| Serpin peptidase inhibitor clade B (ovalbumin), member 9 | BC002538 | SERPINB9 | 2.59 | Anti-apoptosis signal transduction |
| Collagen, type VI, alpha 2 | NM_001849 | COL6A2 | 2.53 | Organization and biogenesis |
| Family with sequence similarity 108 member 1 | NM_031213 | FAM108A1 | 2.49 | Hydrolase activity |
| WW, C2 and coiled-coil domain containing 1 | NM_015238 | WWC1 | 2.46 | Cellular function |
| ADP-ribosylation factor 3 | NM_001659 | ARF3 | 2.45 | GTPase signal transduction |
| ATPase, Class V, type 10C | NM_024490 | ATP10A | 2.45 | Cation transport |
| Immunoglobulin heavy constant mu | BC001872 | IGHG1 | 2.43 | Immune response |
| Immediate early response 3 interacting protein 1 | NM_016097 | IER3IP1 | 2.33 | Integral to membrane |
| ATP-binding cassette (CFTR/MRP), member 10 | NM_033450 | ABCC10 | 2.3 | Transport |
| Rabaptin, RAB GTPase binding effector protein 2 | NM_024816 | RABEP2 | 2.27 | Endocytosis, protein transport |
| Essential meiotic endonuclease 1 homolog 1 | NM_152463 | EME1 | 2.27 | DNA repair |
| Misshapen-like kinase 1(zebrafish) (MINK1 | NM_015716 | MINK1 | 2.26 | Protein Phosphorylation |
| Arachidonate 5-lipoxygenase | NM_000698 | ALOX5 | 2.25 | Linoleic acid metabolism |
| Secreted phosphoprotein 1 | NM_001040058 | SPP1 | 2.18 | TGF Beta Signaling Pathway |
| Small nuclear ribonucleoprotein polypeptide A | NM_004596 | SNRPA | 2.16 | mRNA processing |
| Member RAS oncogene family | NM_021168 | RAB40C | 2.15 | GTPase signal transduction |
| RUN and TBC1 domain containing 1 | BC029251 | RUTBC1 | 2.13 | Unknown |
| Protease, serine, 21 | NM_006799 | PRSS21 | 2.13 | Proteolysis |
| Additional sex combs like 1 | NM_015338 | ASXL1 | 2.1 | Transcription regulation |
| Insulin-like growth factor 2 mRNA binding protein 2 | NM_006548 | IGF2BP2 | 2.08 | Protein biosynthesis |
| Zinc finger protein 432 | NM_014650 | ZNF432 | 2.07 | Transcription regulation |
| Metastasis associated lung adenocarcinoma transcript 1 | NR_002819 | MALAT1 | 2.07 | Binding |
| UDP-N-acetyl-alpha-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase-like 1 | AI097463 | GALNTL1 | 2 | Glycan biosynthesis |
| Higher Expression in Male | ||||
|---|---|---|---|---|
| Gene Name | Accession No. | Gene ID | Ratio | Ontology |
| Ribosomal protein S4, Y-linked (RPS4Y) | NM_001008 | RPS4Y1 | 29.65 | Protein Biosysnthesis |
| DEADH (Asp-Glu-Ala-AspHis) box polypeptide, Y linked | NM_004660 | DDX3Y | 16.17 | Nucleotide binding |
| Chromosome Y open reading frame 15A, Testis protein | AF332224 | CYorf15A | 15.48 | |
| Eukaryotic translation initiation factor 1A, Y-linked | BC005248 | EIF1AY | 14.74 | Protein Biosysnthesis |
| Smcy homolog, Y-linked | NM_004653 | SMCY | 11.81 | Transcription regulation |
| Zinc finger protein, Y-linked | NM_003411 | ZFY | 6.79 | Transcription regulation |
| Ubiquitin specific peptidase 9, Y-linked | NM_004654 | USP9Y | 5.28 | Ubiquitin-dependent protein catabolism |
| Lumican | NM_002345 | LUM | 2.82 | visual perception, Collagen fibril organization |
| Cofilin 1(non-muscle) | NM_005507 | CFL1 | 2.63 | Signal Transduction |
| SH3 domain binding glutamic acid-rich protein like | NM_003022 | SH3BGRL | 2.61 | SH3/SH2 adaptor activity |
| Cysteine-rich secretory protein 2 | NM_003296 | CRISP2 | 2.51 | Testis Specific |
| Acid phosphatase, testicular | NM_033068 | ACPT | 2.43 | Riboflavin metabolism |
| RNA binding motif protein 3 | NM_006743 | RBM3 | 2.33 | RNA Processing |
| Rho GTPase activating protein 5 | NM_001173 | ARHGAP5 | 2.24 | GTPase mediated signal transduction |
| Inhibin beta A | NM_002192 | INHBA | 2.22 | TGF Beta Signaling Pathway |
| Short coiled-coil protein | NM_032547 | SCOC | 2.18 | Unknown |
| Crystallin, beta B2 | NM_000496 | CRYBB2 | 2.16 | Visual perception |
| Apolipoprotein D | NM_001647 | APOD | 2.15 | Lipid metabolism |
| Similar to ubiquitin B precursor | NM_018955 | UBB | 2.13 | Protein ubiquitination |
| HBS1-like | NM_006620 | HBS1L | 2.12 | Protein Biosynthesis |
| Replication protein A3 | NM_002947 | RPA3 | 2.11 | DNA replication |
| Chromosome 1 open reading frame 43 | NM_015449 | C1orf43 | 2.11 | Unknown |
| Cysteine/tyrosine-rich 1 | NM_052954 | CYYR1 | 2.1 | Unknown |
| Basic helix-loop-helix domain containing, class B, 2 | NM_003670 | BHLHB2 | 2.08 | Transcription regulation |
| Chromosome 1 open reading frame 80 | NM_022831 | C1orf80 | 2.06 | Unknown |
| Fatty acid binding protein 7 | NM_001446 | FABP7 | 2.05 | Fatty acid metabolism |
| Ras-related C3 botulinum toxin substrate 1 | NM_006908 | RAC1 | 2.04 | GTPase mediated signal transduction |
Genes listed had a signal intensity of >5.0 in at least one group, expression ratio of >2.0 (between glands), p value <0.05, with known gene identity.
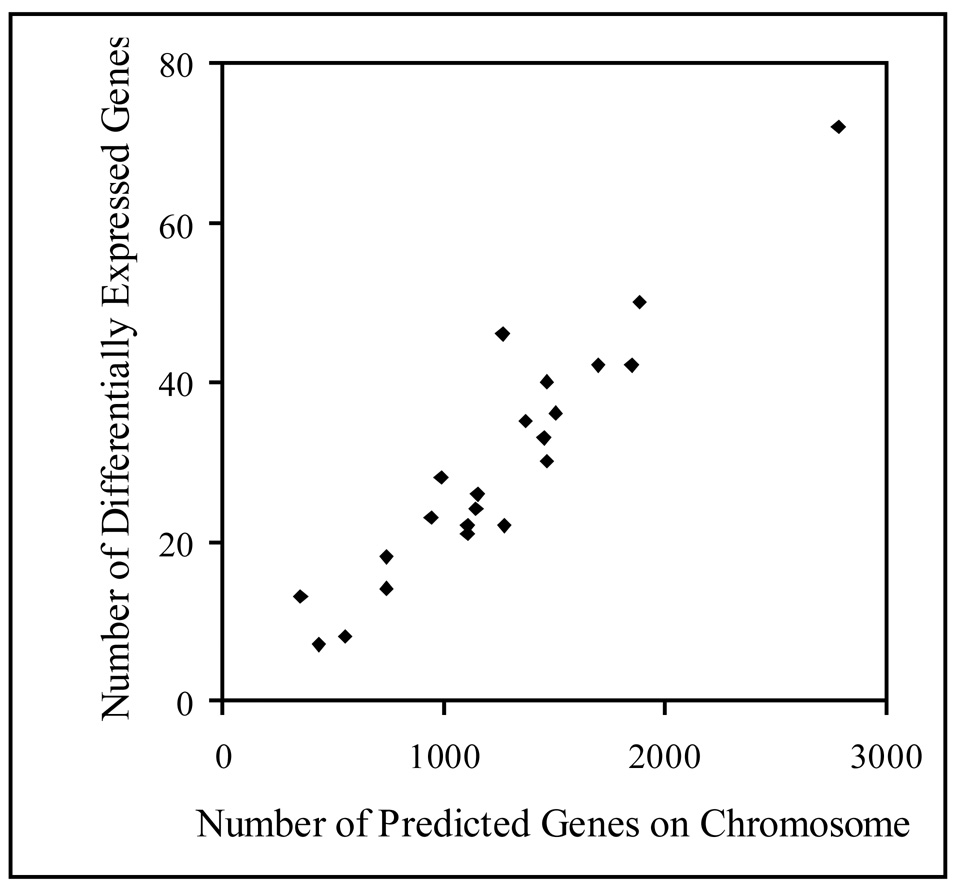
The distribution on chromosomes 1 to 22 of the 652 genes differentially expressed in male and female parotid glands was analyzed, excluding the X and Y sex chromosomes. The number of differentially expressed genes on a given chromosome was directly related to the total number of predicted genes present on that chromosome (Figure 4). This result suggests that the distribution of differentially expressed genes is randomly dispersed throughout the genome.
Figure 4.
A direct correlation in between number of genes on a chromosome and the number of differentially expressed genes on that chromosome
The effect of gender on gene expression in the human parotid gland involved a diverse range of biological processes, molecular functions and cellular components. The z-score analysis indicates the involvement of these genes in several important pathways. As shown in Table 7, gender influenced many genes that affect metabolism, transcription, DNA binding, metal binding and secretory pathway, and they are localized in different cell compartments. Among the differentially expressed genes, several are involved in transcription regulation; e.g., PNN, ASXL1 and ZNF432 are more highly expressed in females, whereas SMCY, ZFY and BHLHB2 are more highly expressed in male parotid tissue (Table 6). Male parotid glands highly expressed submaxillary gland androgen regulated protein B (SMR3A, ont = secretion) and apolipoprotein D (APOD, ont = lipid metabolism); whereas, genes highly expressed in female glands were SPDI, transmembrane family member 2 (SID2, ont = lipid metabolism) and cytochrome B5 reductase (CYB5R3, ont = electron transport) (see GEO # GSE8764). (Supplement Table B).
Table 7.
Gene expression in male and female human parotid glands and their ontological categorization
| Ontology | Diff | M | F | Total | M | F |
|---|---|---|---|---|---|---|
| exp genesa | upb | upc | on Arrayd | z-scoree | z-scoref | |
| Biological Processes | ||||||
| Physiological process | 387 | 176 | 211 | 9550 | 1.54 | 2.21 |
| Metabolism | 275 | 122 | 153 | 6530 | 0.99 | 2.25 |
| Regulation of biological process | 139 | 51 | 88 | 3375 | −1.34 | 2.53 |
| Protein metabolism | 122 | 64 | 58 | 2623 | 3.01 | 0.51 |
| Transcription | 76 | 23 | 53 | 1977 | −2.24 | 2.04 |
| Cell organization and biogenesis | 66 | 38 | 28 | 1405 | 2.86 | −0.26 |
| Protein transport | 33 | 16 | 17 | 534 | 2.22 | 1.82 |
| Protein biosynthesis | 28 | 17 | 11 | 525 | 2.63 | 0.01 |
| Vesicle-mediated transport | 25 | 15 | 10 | 352 | 3.62 | 1.01 |
| Protein kinase cascade | 19 | 10 | 9 | 278 | 2.35 | 1.36 |
| Response to wounding | 19 | 5 | 14 | 363 | −0.57 | 2.4 |
| Macromolecule catabolism | 18 | 11 | 7 | 332 | 2.18 | 0.03 |
| Secretion | 16 | 12 | 4 | 250 | 3.69 | −0.54 |
| Endocytosis | 12 | 6 | 6 | 130 | 2.48 | 2.03 |
| Cellular Components | ||||||
| Intracellular | 329 | 139 | 190 | 7016 | 1.78 | 4.34 |
| Organelle | 259 | 108 | 151 | 5757 | 0.55 | 2.8 |
| Membrane-bound organelle | 231 | 97 | 134 | 5065 | 0.77 | 2.62 |
| Cytoplasm | 161 | 82 | 79 | 3131 | 4.01 | 1.25 |
| Nucleus | 155 | 56 | 99 | 3468 | −1.04 | 2.93 |
| Integral to membrane | 117 | 54 | 63 | 3457 | −1.32 | −2.02 |
| Protein complex | 78 | 41 | 37 | 1569 | 2.58 | 0.33 |
| Mitochondrion | 34 | 21 | 13 | 618 | 3.05 | −0.24 |
| Endoplasmic reticulum | 31 | 19 | 12 | 579 | 2.73 | −0.29 |
| Organelle membrane | 30 | 15 | 15 | 474 | 2.26 | 1.38 |
| Extracellular region | 28 | 15 | 13 | 998 | −0.76 | −2.11 |
| Ribonucleoprotein complex | 23 | 18 | 5 | 330 | 5.04 | −0.91 |
| Envelope | 19 | 10 | 9 | 252 | 2.6 | 1.44 |
| Molecular Processes | ||||||
| Protein binding | 204 | 97 | 107 | 4621 | 2.51 | 0.77 |
| Ion binding | 120 | 41 | 79 | 3370 | −2.64 | 0.74 |
| Nucleotide binding | 99 | 39 | 60 | 1790 | 1.61 | 3.65 |
| Transferase activity | 70 | 22 | 48 | 1665 | −1.35 | 2.1 |
| ATP coupled activity and binding | 60 | 22 | 38 | 1196 | 0.33 | 2.47 |
| Kinase activity | 37 | 11 | 26 | 733 | −0.47 | 2.6 |
| Pyrophosphatase activity | 33 | 19 | 14 | 502 | 3.63 | 0.95 |
| GTP binding | 23 | 14 | 9 | 290 | 4.12 | 1.08 |
| Transcription factor activity | 23 | 6 | 17 | 791 | −2.15 | −0.07 |
| Structural constituent of ribosome | 12 | 9 | 3 | 150 | 4.06 | −0.16 |
| Calmodulin binding | 11 | 3 | 8 | 120 | 0.66 | 3.37 |
| Hydrogen ion transporter activity | 10 | 4 | 6 | 96 | 1.85 | 2.74 |
Diff exp genes indicate the total number of genes differentially expressed on the array in that category.
up regulated genes in male parotid glands, as compared to those of female (M-up).
Up regulated genes in female parotid glands, as compared to those of male (F-up).
'Total on array indicates the total number of genes on the array in that Ontological category.
z-score-M and z-score-F indicate the z-score for the male and female respectively.
The z-scores with value >2.0 or <-2.0 are reported for ontologies with more than 10 differentially expressed genes.
The Affymetrix® GeneChip® HGU133 Plus 2.0 array contains about 250 probe sets related to the secretory pathway, 16 of which were differentially expressed by gender with a z-score of 3.69 (Table 7). Several proteins involved in exocytosis were found to be differentially expressed in parotid tissue, e.g., members of the SNARE complex such as Syntaxin, VAMP, SNAP and proteins involved in the regulation and formation of the SNARE complex, e.g., RAB (Table 8). ARF3, which encodes for a small guanine nucleotide-binding protein that plays a role in vascular trafficking and as an activator of phospholipase D, also showed differential expression. The GTP binding protein SARA showed higher expression in female parotid tissue. Several members of calcium signaling pathways were also differentially expressed, e.g. CAMK2G, Inositol 3 phosphate 3 kinase B (ITPKB), nitric oxide synthase 3 (NOS3), and the plasma membrane calcium ATPase type 2 (PMCA2) (Table 9).
Table 8.
Differentially expressed genes in male and female human parotid glands known to be involved in secretion
| Gene Name | Accession No. | Gene ID | Ratio (M/F) | Direction |
|---|---|---|---|---|
| ADP-ribosylation factor 3 | NM_001659 | ARF3 | 2.45 | Down |
| Calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma | NM_172173 | CAMK2G | 2.38 | Down |
| Inhibin, beta A | NM_002192 | INHBA | 2.22 | Up |
| Signal recognition particle 54kDa | NM_003136 | SRP54 | 1.82 | Up |
| Vesicle-associated membrane protein 3 (cellubrevin) | NM_004781 | VAMP3 | 1.82 | Up |
| RAB1A, member RAS oncogene family | NM_004161 | RAB1A | 1.76 | Up |
| tumor necrosis factor (ligand) superfamily, member 13b | NM_006573 | TNFSF13B | 1.75 | Down |
| RAB1A, member RAS oncogene family | NM_004161 | RAB1A | 1.73 | Up |
| Epidermal growth factor receptor pathway substrate 15-like 1 | NM_021235 | EPS15R | 1.66 | Down |
| SEC24 related gene family, member D | NM_014822 | SEC24D | 1.63 | Up |
| Syntaxin binding protein 3 | NM_007269 | STXBP3 | 1.63 | Up |
| Caspase recruitment domain family, member 8 | NM_014959 | CARD8 | 1.61 | Down |
| Translocation associated membrane protein 1 | NM_014294 | TRAM1 | 1.61 | Up |
| RAB2, member RAS oncogene family | NM_002865 | RAB2 | 1.61 | Up |
| RAB22A, member RAS oncogene family | NM_020673 | RAB22A | 1.61 | Up |
| Folate receptor 1 | NM_016731 | FOLR1 | 1.59 | Up |
| GTP-binding protein Sara | NM_016103 | SAR1B | 1.59 | Up |
| ATP-binding cassette, sub-family A (ABC1), member 1 | NM_005502 | ABCA1 | 1.58 | Down |
| Syntaxin binding protein 1 | NM_003165 | STXBP1 | 1.56 | Down |
| Synaptosomal-associated protein, 23kDa | NM_130798 | SNAP23 | 1.53 | Up |
Table 9.
Differentially expressed genes in male and female parotid glands known to be involved in calcium signaling pathways
| Gene Name | Accession No. | Gene ID | Ratio (M/F) | direction |
|---|---|---|---|---|
| Inositol 1,4,5-triphosphate 3-kinase B | NM_002221 | IP3K | 1.8 | down |
| Nitric Oxide synthase 3 | NM_024711 | NOS3 | 1.65 | down |
| Calcium clamoduline-dependent protein kinase (CaM Kinase) II gamma | NM_172170 | CAMK2G | 2.38 | down |
| ATPase, Ca2+ transporting, plasma membrane | X63575 | PMCA2 | 1.72 | down |
| Voltage-dependent anion channel 3 | U90943 | VDAC3 | 1.77 | up |
| Guanine nucleotide binding protein (G protein) q polypeptide | NM_002072 | GNAQ | 1.98 | up |
Validation of Array Data
Gender Related Differences
In addition to the RNA samples analyzed by microarray, RNA samples from another 19 subjects were also isolated for Q-PCR analysis to validate the gender-specific array results. This second group included 11 female subjects (27, 35, 36, 40, 43, 49, 53, 61, 66, 74 and 83 years of age) and 8 males (40, 46, 51, 55, 67, 70, 73 and 85 years old). Four genes which were differentially expressed in human parotid glands were selected for Q-PCR evaluation. Genes highly expressed in male glands included DDX3Y, ZFY and CYTORF15, whereas a gene highly expressed in female glands was XIST. Q-PCR results confirmed the array data (Table 10). Each of the genes tested by Q-PCR showed the same pattern of expression as measured by array technique.
Table 10.
Q-PCR confirmation of selected HGU133A 2.0 microarray results
| Sex Differences (Sample number used for QPCR, Female n= 11; Male n=8) | ||||||
|---|---|---|---|---|---|---|
| AccessioN No. | GeneID | Fold Changes (M/F) | Gene Name | |||
| Microarray | p Value | Q-PCR | p Value | |||
| NR_001564 | XIST | −124.03 | 0.00 | −106.0 | 0.00 | X (inactive)-specific transcript |
| NM_004660 | DDX3Y | 16.17 | 0.00 | 40.0 | 0.00 | DEADH (Asp-Glu-Ala-AspHis) box polypeptide, Y chromosome |
| BC033974 | ZFY | 6.79 | 0.00 | 15.0 | 0.00 | Zinc finger protein, Y-linked |
| AF332225 | CYorf15B | 15.48 | 0.00 | 22.0 | 0.00 | Chromosome Y open reading frame 15B |
| Differences due to aging (Sample number used for QPCR, Young n = 8; Old n = 8) | ||||||
| Accession No. | GeneID | Fold Changes (O/Y) | Gene Name | |||
| Microarray | p Value | Q-PCR | p Value | |||
| BC002704 | STAT1 | −3.3 | 0.035 | −1.51 | 0.03 | Signal transducer and activator of transcription 1 |
| NM_001565 | CXCL10 | −8.37 | 0.041 | −1.96 | 0.05 | Small inducible cytokine subfamily B (Cys-X-Cys), member 10 |
| NM_002122 | HLA-DQA1 | −3.5 | 0.035 | −1.57 | 0.05 | Histocompatibility complex, class II, DQ alpha 1 |
| NM_152679 | SLC10A4* | 4.41 | 0.032 | 2.00 | 0.05 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
| NM_138455 | CTHRC1* | 2.82 | 0.047 | 1.81 | 0.05 | Collagen triple helix repeat containing 1 |
| AI500293 | CHRM1* | −1.61 | 0 | −1.28 | 0.04 | Cholinergic receptor, muscarinic 1 |
Tested with mixed population of male and female young and old subjects
Age Related Differences
To verify the differential gene expression in the young and old female populations as observed by microarray analysis, three genes were selected for Q-PCR (STAT1, CXCL10 and HLA-DQA1). The female parotid from 8 young (19, 27, 35, 36, 38, 40, 43 and 49 years old) and 8 older (53, 61, 65, 65, 66, 69, 74 and 83 years) subjects were used for this analysis. Table 10 shows that the results of Q-PCR study gave the same gene expression pattern as that obtained by microarray. Three genes (SLC10A4, CTHRC1 and CHRM1) were further tested with mixed gender populations of young (19, 27, 36, 38 and 49 year old females; and 40, 46 and 51 year old males) and old (61, 66, 74 and 83 year old females; and 67, 70, 73 and 85 year old males) subjects. As shown in Table 10, all three genes selected for validation by Q-PCR technique further confirmed the array results.
DISCUSSION
Although well documented in rodents, this is the first comprehensive report to demonstrate the inherent gender-specific differences in gene expression in human parotid gland. Our findings are consistent with the gender associated and gland specific variations in mRNA levels previously reported in rodent model salivary gland systems (20, 27, 28) and in lacrimal (29) and meibomian glands (30). The differentially expressed genes in these studies are involved in a wide range of biological processes, molecular functions and cellular components, including growth and development, transcription, metabolism, signal transduction, ion transport, receptor activity and protein and nucleic acid binding. In the present study, gender-specific differences in expression were noted for 787 out of 54,000 probe sets on the HG U133 Plus 2.0 array, with the majority of these genes being expressed to a higher extend in females (~59%). The proteins encoded by these genes are located in different cell compartments, i.e. the nucleus, plasma membrane, mitochondria and cytoplasm. At this time, we can only speculate as to the biological and physiological implications of the observed sexual dimorphism in salivary gland gene expression. It should be noted that the sexual dimorphism in mice is in part due to gland-specific differences in gene expression between males and females (20), consistent with the gender-related differences in human salivary glands being due to tissue-specific variations in gene expression.
Gender-specific differential gene expression was detected on all chromosomes and the number of differences was found to be directly related to the size of the chromosome, i.e. the larger the chromosome, the greater the number of differences in gene expression that were detected (a similar chromosomal distribution was detected for aging, not shown) suggesting that the distribution of differentially expressed genes is randomly dispersed throughout the genome. The differential gene expression pattern on the X and Y chromosomes of several genes (e.g. UTX, DDX3X, SMCX) are in agreement with previous reports on human lympholastoma cell line (31) and 11 different human tissues (32). The gene ontologies analysis of gender-specific, differential expression patterns provides several examples of genes that potentially explain how gender modulates salivation by the human parotid gland (1, 4, 15).
Age related differences in gene expression have been noted in the mouse submandibular gland model (33). Using cDNA array analysis, Hiratsuka et al. found that 160 of the 1328 genes screened showed more than a two-fold change, 96% of which exhibited decreased expression in elderly mice (33). These genes are associated with numerous biological pathways, e.g., transcription regulation, ion transport, and signal transduction. The effects of age on specific gene ontologies in the human parotid gland may provide insight into functional and morphological changes previously described (2, 3, 16).
We also found that age had a significant influence on the expression of genes associated with primary metabolism and physiological processes. These observations were not unexpected considering that aged animals demonstrate a reduced protein biosynthesis, but importantly, our results provide novel information defining which specific genes may be most affected by aging. Of particular interest are those associated with defense/immune responses. The expression of both HLA-DQA1 and HLA-DQB1 are decreased in the parotid gland of the aged. HLA-DQA1 and HLA-DQB1 belong to the histocompatibility complex loci (HLA) class II. The class II molecule is a heterodimer consisting of an alpha (DQA) and beta (DQB) chain, both anchored in the membrane. HLA II plays a central role in the immune system by T-cell activation (34, 35). Chemokine (C-X-C motif) ligand 10 (CXCL10) also showed lower expression in the aged population. Chemokines are a group of low molecular weight peptides that induce the chemotaxis of different leukocyte subtypes. At present, more than 50 chemokines have been described. CXC chemokines attract neutrophils and promote their adherence to endothelial cells. Several other proteins known to be involved in different immune response pathways showed altered expression in aged population (e.g., IRF1, IRF7, GBP1, IFITM1, IFITM2, IFITM3, PSMB8 and PSMB9). Complex remodeling of the immune system occurs during aging, which may contribute significantly to morbidity and mortality in the elderly (36, 37). Despite the great number of studies on changes in the immune system of the elderly, the biological basis of such changes is unclear. This is at least partly due to the alterations observed in the immune system of the elderly that could be a cause or the consequence of the underlying pathological processes. Undoubtedly, diseases such as infectious, autoimmune and neoplastic pathologies, which aged people are particularly susceptible to, involve dysregulation of immune function (36, 37). On the other hand, recent studies in healthy centenarians suggest that the immunological changes observed during aging are consistent with a reshaping, rather than a generalized deterioration, of the main immune functions (38).
The number of elderly is dramatically increasing, and consequently, geriatric pathology is becoming a more important aspect of clinical practice. Therefore, it is particularly important to evaluate further the findings in the immune system of the elderly so as to better understand their susceptibility to certain diseases, and the links between health and longevity. Salivary gland function may prove to be a parameter worth evaluating in the aged, as shown in other clinical populations (39,40).
This study is an initial important step in identifying the genes which are differentially expressed due to gender and aging. It is noteworthy that very little overlap was observed between the gender-related and age-related differences in gene expression (<1.3%), indicating that these differences are specific. Our results will hopefully stimulate additional studies in this area, especially clinical studies that aid in the development of strategies to reverse or lessen the negative impact of age-related changes in gene expression on oral health. Although the amount of data obtained from microarray can be overwhelming, informatics tools are emerging that take high quality datasets and permit systems level analysis that can identify key biological pathways and genes that are involved in normal physiological and pathogenic processes (41). Q-PCR analysis confirmed the results of the microarray study, and verified reproducibility of our results in additional independent samples, indicating that glands from another population of subjects of the same age range and/or gender group would very likely generate the same results. Therefore, our results provide critical information for understanding the complex changes in gene expression that may significantly contribute to gender-associated and age-related differences in the secretion mechanism.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Zugen Chen, Jason Liu and Shilpa Patel at UCLA microarray core facility for their cooperation in generating the microarray data. We also thank Eastern, Mid Western and South Division of the Cooperative Human Tissue Network for the supply of the human salivary gland tissue. This research was supported in part by PHS grants RO1-DE09692, R37-DE08921 (JEM), T32-DE07202 (AS), RO1-DE17593 (DTW) and T32-DE07296 (JW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Percival RS, Challacombe SJ, Marsh PD. Flow rates of resting whole and stimulated parotid saliva in relation to age and gender. J Dent Res. 1994;73(8):1416–1420. doi: 10.1177/00220345940730080401. [DOI] [PubMed] [Google Scholar]
- 2.Yeh CK, Johnson DA, Dodds MW. Impact of aging on human salivary gland function: a community-based study. Aging (Milano) 1998;10(5):421–428. doi: 10.1007/BF03339889. [DOI] [PubMed] [Google Scholar]
- 3.Eliasson L, Birkhed D, Osterberg T, Carlen A. Minor salivary gland secretion rates and immunoglobulin A in adults and the elderly. Eur J Oral Sci. 2006;114(6):494–499. doi: 10.1111/j.1600-0722.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- 4.Heintze U, Birkhed D, Bjorn H. Secretion rate and buffer effect of resting and stimulated whole saliva as a function of age and sex. Swed Dent J. 1983;7(6):227–238. [PubMed] [Google Scholar]
- 5.Pedersen W, Schubert M, Izutsu K, Mersai T, Truelove E. Age-dependent decreases in human submandibular gland flow rates as measured under resting and post-stimulation conditions. J Dent Res. 1985;64(5):822–825. doi: 10.1177/00220345850640050801. [DOI] [PubMed] [Google Scholar]
- 6.Tylenda CA, Ship JA, Fox PC, Baum BJ. Evaluation of submandibular salivary flow rate in different age groups. J Dent Res. 1988;67(9):1225–1228. doi: 10.1177/00220345880670091501. [DOI] [PubMed] [Google Scholar]
- 7.Wu AJ, Baum BJ, Ship JA. Extended stimulated parotid and submandibular secretion in a healthy young and old population. J Gerontol A Biol Sci Med Sci. 1995;50A(1):M45–M48. doi: 10.1093/gerona/50a.1.m45. [DOI] [PubMed] [Google Scholar]
- 8.Fischer D, Ship JA. Effect of age on variability of parotid salivary gland flow rates over time. Age Ageing. 1999;28(6):557–561. doi: 10.1093/ageing/28.6.557. [DOI] [PubMed] [Google Scholar]
- 9.Elishoov H, Wolff A, Volovikov A, Gorsky M. [Evaluation of unstimulated and stimulated parotid salivary flow rate in Israeli healthy subjects aged 60 years and older] Refuat Hapeh Vehashinayim. 2005;22(2):44–48. 86. [PubMed] [Google Scholar]
- 10.Cook D, Van Lennep EW, Roberts ML, Young JA. Secretion by the Major Salivary Glands. In: Johnson LR, editor. Physiology of the Gastrointestinal Tract. New York: Raven Press; 1994. pp. 1061–1117. [Google Scholar]
- 11.Turner RJ, Sugiya H. Understanding salivary fluid and protein secretion. Oral Dis. 2002;8(1):3–11. doi: 10.1034/j.1601-0825.2002.10815.x. [DOI] [PubMed] [Google Scholar]
- 12.Guo T, Rudnick PA, Wang W, Lee CS, Devoe DL, Balgley BM. Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J Proteome Res. 2006;5(6):1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 13.Melvin JE, Yule D, Shuttleworth T, Begenisich T. Regulation of fluid and electrolyte secretion in salivary gland acinar cells. Annu Rev Physiol. 2005;67:445–469. doi: 10.1146/annurev.physiol.67.041703.084745. [DOI] [PubMed] [Google Scholar]
- 14.Ship JA. Diagnosing, managing, and preventing salivary gland disorders. Oral Dis. 2002;8(2):77–89. doi: 10.1034/j.1601-0825.2002.2o837.x. [DOI] [PubMed] [Google Scholar]
- 15.Inoue H, Ono K, Masuda W, Morimoto Y, Tanaka T, Yokota M, et al. Gender difference in unstimulated whole saliva flow rate and salivary gland sizes. Arch Oral Biol. 2006;51(12):1055–1060. doi: 10.1016/j.archoralbio.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Vissink A, Spijkervet FK, Van Nieuw Amerongen A. Aging and saliva: a review of the literature. Spec Care Dentist. 1996;16(3):95–103. doi: 10.1111/j.1754-4505.1996.tb00842.x. [DOI] [PubMed] [Google Scholar]
- 17.Brown DA, Bruce JI, Straub SV, Yule DI. cAMP potentiates ATP-evoked calcium signaling in human parotid acinar cells. J Biol Chem. 2004;279(38):39485–39494. doi: 10.1074/jbc.M406201200. [DOI] [PubMed] [Google Scholar]
- 18.Nakamoto T, Srivastava A, Romanenko VG, Ovitt CE, Perez-Cornejo P, Arreola J, et al. Functional and molecular characterization of the fluid secretion mechanism in human parotid acinar cells. Am J Physiol Regul Integr Comp Physiol. 2007;292(6):R2380–R2390. doi: 10.1152/ajpregu.00591.2006. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treister NS, Richards SM, Lombardi MJ, Rowley P, Jensen RV, Sullivan DA. Sex-related differences in gene expression in salivary glands of BALB/c mice. J Dent Res. 2005;84(2):160–165. doi: 10.1177/154405910508400210. [DOI] [PubMed] [Google Scholar]
- 21.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Robinson JF, Khan HM, Carter DE, McKinney J, Miskie BA, et al. Optimizing RNA extraction yield from whole blood for microarray gene expression analysis. Clin Biochem. 2004;37(9):741–744. doi: 10.1016/j.clinbiochem.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Thach DC, Shaffer KM, Ma W, Stenger DA. Assessing the feasibility of using neural precursor cells and peripheral blood mononuclear cells for detection of bioactive Sindbis virus. Biosens Bioelectron. 2003;18(8):1065–1072. doi: 10.1016/s0956-5663(02)00246-4. [DOI] [PubMed] [Google Scholar]
- 27.Murphy RA, Watson AY, Metz J, Forssmann WG. The mouse submandibular gland: an exocrine organ for growth factors. J Histochem Cytochem. 1980;28(8):890–902. doi: 10.1177/28.8.6969274. [DOI] [PubMed] [Google Scholar]
- 28.Senorale-Pose M, Jacqueson A, Rougeon F, Rosinski-Chupin I. Acinar cells are target cells for androgens in mouse submandibular glands. J Histochem Cytochem. 1998;46(5):669–678. doi: 10.1177/002215549804600512. [DOI] [PubMed] [Google Scholar]
- 29.Richards SM, Jensen RV, Liu M, Sullivan BD, Lombardi MJ, Rowley P, et al. Influence of sex on gene expression in the mouse lacrimal gland. Exp Eye Res. 2006;82(1):13–23. doi: 10.1016/j.exer.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Richards SM, Yamagami H, Schirra F, Suzuki T, Jensen RV, Sullivan DA. Sex-related effect on gene expression in the mouse meibomian gland. Curr Eye Res. 2006;31(2):119–128. doi: 10.1080/02713680500514644. [DOI] [PubMed] [Google Scholar]
- 31.McRae AF, Matigian NA, Vadlamudi L, Mulley JC, Mowry B, Martin NG, et al. Replicated effects of sex and genotype on gene expression in human lymphoblastoid cell lines. Hum Mol Genet. 2007;16(4):364–373. doi: 10.1093/hmg/ddl456. [DOI] [PubMed] [Google Scholar]
- 32.Talebizadeh Z, Simon SD, Butler MG. X chromosome gene expression in human tissues: male and female comparisons. Genomics. 2006;88(6):675–681. doi: 10.1016/j.ygeno.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiratsuka K, Kamino Y, Nagata T, Takahashi Y, Asai S, Ishikawa K, et al. Microarray analysis of gene expression changes in aging in mouse submandibular gland. J Dent Res. 2002;81(10):679–682. doi: 10.1177/154405910208101005. [DOI] [PubMed] [Google Scholar]
- 34.Zang W, Murphy B. Peptide-mediated immunosuppression. Am J Ther. 2005;12(6):592–599. doi: 10.1097/01.mjt.0000178766.60234.e2. [DOI] [PubMed] [Google Scholar]
- 35.O'Hanlon TP, Carrick DM, Targoff IN, Arnett FC, Reveille JD, Carrington M, et al. Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: distinct HLA-A, -B, -Cw, -DRB1, and -DQA1 allelic profiles distinguish European American patients with different myositis autoantibodies. Medicine (Baltimore) 2006;85(2):111–127. doi: 10.1097/01.md.0000217525.82287.eb. [DOI] [PubMed] [Google Scholar]
- 36.Katz JM, Plowden J, Renshaw-Hoelscher M, Lu X, Tumpey TM, Sambhara S. Immunity to influenza: the challenges of protecting an aging population. Immunol Res. 2004;29(1–3):113–124. doi: 10.1385/IR:29:1-3:113. [DOI] [PubMed] [Google Scholar]
- 37.Vesosky B, Turner J. The influence of age on immunity to infection with Mycobacterium tuberculosis. Immunol Rev. 2005;205:229–243. doi: 10.1111/j.0105-2896.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 38.Franceschi C, Bonafè M. Centenarians as a model for healthy aging. Biochem Soc Trans. 2003;31:457–461. doi: 10.1042/bst0310457. [DOI] [PubMed] [Google Scholar]
- 39.Nieuw Amerongen AV, Ligtenberg AJ, Veerman EC. Implications for diagnostics in the biochemistry and physiology of saliva. Ann N Y Acad Sci. 2007;1098:1–6. doi: 10.1196/annals.1384.033. [DOI] [PubMed] [Google Scholar]
- 40.Samaranayake L. Saliva as a diagnostic fluid. Int Dent J. 2007;57(5):295–299. doi: 10.1111/j.1875-595x.2007.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1(1):54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.