Abstract
This review deals with biochemical and physiological aspects of plant ornithine d-aminotransferase (OAT, EC 2.6.1.13). OAT is a mitochondrial enzyme containing pyridoxal-5′-phosphate as a cofactor, which catalyzes the conversion of L-ornithine to L-glutamate γ-semialdehyde using 2-oxoglutarate as a terminal amino group acceptor. It has been described in humans, animals, insects, plants and microorganisms. Based on the crystal structure of human OAT, both substrate binding and reaction mechanism of the enzyme are well understood. OAT shows a large structural and mechanistic similarity to other enzymes from the subgroup III of aminotransferases, which transfer an amino group from a carbon atom that does not carry a carboxyl function. In plants, the enzyme has been implicated in proline biosynthesis and accumulation (via pyrroline-5-carboxylate), which represents a way to regulate cellular osmolarity in response to osmotic stress. However, the exact metabolic pathway involving OAT remains a subject of controversy.
Key words: ornithine δ-aminotransferase, osmotic stress, proline, Δ1-pyrroline-5-carboxylate, pyridoxal-5′-phosphate, semialdehyde, transamination
Introduction
Ornithine δ-aminotransferase or 5-aminotransferase (OAT; L-ornithine:2-oxoacid aminotransferase; EC 2.6.1.13) is a nuclearencoded, pyridoxal-5′-phosphate (PLP)-dependent enzyme found in the mitochondrial matrix of most human and animal tissues.1 The enzyme also exists in insects,2 microorganisms3 and plants.4 It catalyzes the transfer of the δ-amino group of L-ornithine to 2-oxoglutarate, which produces L-glutamate γ-semialdehyde (GSA) and L-glutamate. Some enzymes can utilize pyruvate or glyoxylate as amino group acceptors instead of 2-oxoglutarate.3,5 N-acetylornithine represents an alternative substrate for OAT from Plasmodium falciparum.6 Human OAT is expressed as a precursor, which is processed into its mature form following the cleavage of the mitochondrial transit peptide upon entry to the mitochondrion.7 Putative signal peptides occur also in sequences of plant OAT precursors.4,8 The mitochondrial localization of OAT in Arabidopsis was demonstrated using a green fluorescent protein-based reporter fusion.9
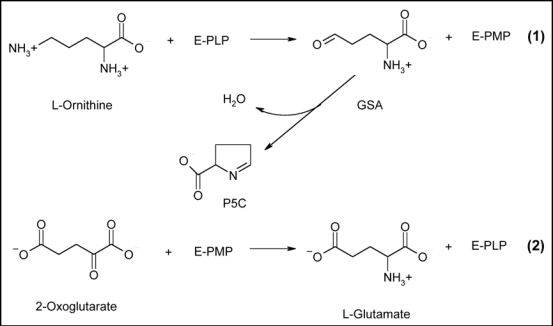
Similar to all PLP-dependent aminotransferases, the transamination reaction proceeds via a ping-pong mechanism that requires two half reactions to complete one catalytic cycle of transamination10 (Fig. 1). In the first half reaction, the PLP cofactor forms an aldimine with the δ-amino group of L-ornithine. Then a stereospecific proton transfer occurs from the δ-carbon of L-ornithine to the C4' carbon of PLP. The aldimine (“enzyme-substrate Schiff base”) is converted to another aldimine (“enzyme-product Schiff base”) between pyridoxamine-5′-phosphate (PMP) and GSA. Subsequent hydrolysis releases GSA, which spontaneously cyclizes to form Δ1-pyrroline-5-carboxylate (P5C), and PMP. The first half reaction is virtually irreversible.10 In the second half-reaction, 2-oxoglutarate undergoes transamination to L-glutamate, while the cofactor is restored to its PLP form.10
Figure 1.
The catalytic mechanism of OAT. The presented reaction scheme10 points out the role of the enzyme cofactor PLP. Numbers indicate the two separate half reactions.
Plant Ornithine δ-Aminotransferases
Transamination of L-ornithine in higher plants was demonstrated fifty years ago, namely in spinach (Spinacia oleracea)11 and mungbean (Phaseolus aureus).12 The mungbean enzyme was found to be associated with the mitochondria.12 In the following two decades, OAT was partially purified for example from peanut (Arachis hypogea),13 pumpkin (Cucurbita maxima)14 and squash (Cucurbita pepo).15 By activity measurements in cotyledon extracts, OAT was demonstrated in pea (Pisum sativum).16 For the squash enzyme, the pH optimum of the transamination between L-ornithine and 2-oxoglutarate was 8.0 and the Michaelis constants were 4.7 and 6.3 mM, respectively. Squash OAT showed a molecular mass of 48 kDa as determined by gel chromatography.15
Proline auxotroph mutants of E. coli were transformed by mothbean (Vigna aconitifolia) cDNA expression library, and ornithine and proline prototrophy was restored. This strategy called “trans-complementation” facilitated the isolation of cDNA clones encoding OAT.4 The mothbean enzyme expressed in E. coli was partially purified (a monomer of 50 kDa) and its Km values for ornithine (2 mM) and 2-oxoglutarate (0.75 mM) were determined at an optimal pH of 8.0.4 Recombinant mothbean OAT was found to be inhibited significantly by isoleucine, valine and serine, whereas proline showed no apparent effect (at a concentration of 1 mM of each).17
Ten years ago, a cDNA encoding OAT from Arabidopsis thaliana was obtained and sequenced.8 At present, a number of other plant OAT sequences (e.g., from aquilegia, barrel medic, grape, maize, pine, potato, rice, sorghum and soybean) are available in public DNA and protein databases. The amino acid sequence of pea OAT, UniProtKB accession no. B1A0U3, has been determined in our laboratory (Stránská J. et al., unpublished results) via cloning and sequencing the respective cDNA (EMBL/GenBank accession no. EU414030).
Physiological Role in Plants
L-Arginine is an important storage and transport form of organic nitrogen in many higher plants. For example, developing pea seeds accumulate arginine-rich storage proteins and arginine is the predominant free amino acid in pea cotyledons, accounting for 66.4% of all nitrogen in the amino acid pool of young cotyledons.16 During seed germination, the first steps of arginine degradation are catalyzed by arginase (EC 3.5.3.1), OAT, and urease (EC 3.5.1.5). Arginase hydrolyzes L-arginine to yield urea, which is further degraded by urease to carbon dioxide and ammonia, and L-ornithine providing GSA by OAT reaction. This pathway appears to be related to the transfer of nitrogen from arginine to other amino acids.16 In addition, arginine and ornithine are the precursors of polyamines (putrescine, spermidine and spermine), which are known as compounds with important roles in developmental processes and stress tolerance.18
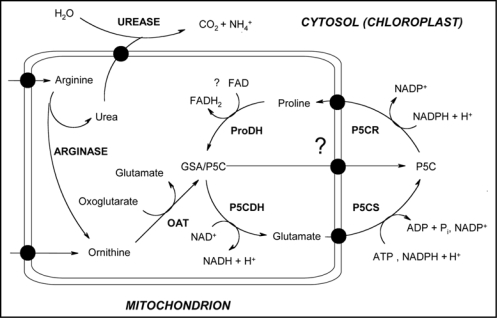
The accumulation of compatible solutes such as polyols/sugars (e.g., mannitol, trehalose), quarternary ammonium compounds (e.g., glycine betaine) and neutral amino acids (e.g., proline) represents one of the most common responses for regulating cellular osmolarity.19 These low-molecular-mass compounds are accumulated to high intracellular concentrations, in order to balance the osmotic pressure of the growth medium and thereby maintain both turgor and the driving gradient for water uptake.19 In plants, proline is mainly synthesized in the cytosol from glutamate via P5C by the sequential action of P5C synthetase (P5CS; a bifunctional enzyme, EC 1.2.1.41/2.7.2.11) and P5C reductase (P5CR; EC 1.5.1.2). For degradation, proline is imported into mitochondria where it is converted back to glutamate by proline dehydrogenase (ProDH; EC 1.5.99.8) and P5C dehydrogenase (P5CDH; EC 1.5.1.12);20–22 Figure 2. There is also evidence for a pathway of proline biosynthesis from ornithine, in which OAT has been implicated.22
Figure 2.
Ivolvement of plant OAT in proline synthesis. The scheme is based on that previously published by Funck D. et al., 2008.9 OAT links the catabolic pathways for arginine and proline, which converge at the intermediate P5C in mitochondria. Proline biosynthesis occurs in the cytosol or, during stress, in plastids. Due to the chemical instability of GSA/P5C, an export from mitochondria seems unlikely but cannot be excluded. Abbreviations are those used throughout the text.
OAT reaction results in GSA and glutamate. The semialdehyde is in spontaneous equilibrium with its cyclic form P5C, a common intermediate in proline metabolism. Formation of GSA/P5C from ornithine was postulated to constitute an alternative pathway of proline synthesis and accumulation.4 The glutamate pathway is thought to be the primary route for proline synthesis in plants during conditions of osmotic stress and nitrogen limitation whereas the ornithine pathway might function under high nitrogen input.4 A study performed on NaCl-treated cotyledons of radish (Raphanus sativus) seedlings using the specific OAT inhibitor gabaculine demonstrated the contribution of the ornithine pathway to proline synthesis.23 In young Arabidopsis thaliana plantlets, free proline content, P5CS mRNA, OAT activity, and OAT mRNA were all increased by salt-stress treatment.8 Moreover, transgenic Nicotiana plumbaginifolia plants overexpressing OAT from Arabidopsis synthesized more proline than the control plants and showed a higher biomass and a higher germination rate under osmotic stress conditions.24 All these data suggest that the ornithine pathway, together with the glutamate pathway, plays an important role in proline accumulation during osmotic stress in plants.
However, targeting OAT to mitochondria would strongly suggest that P5C enters the degradation pathway of proline rather than its biosynthesis. Proline production via ProDH is energetically unfavorable and due to the chemical instability of GSA/P5C, an export of this compound to the cytosol and thus a contribution to proline synthesis appears unlikely.9 The ornithine pathway in rice leaves seems to contribute little, if any, to proline accumulation under water stress condition. When OAT activity decreased by 75% in consequence of a gabaculine inhibitory treatment under water stress conditions, the proline content was reduced only by 20%. Moreover, cycloheximide, a protein synthesis inhibitor, had no effect on OAT activity induced by water stress but significantly reduced proline accumulation.25
A new insight into this field has been introduced by demonstrating that OAT is essential for nitrogen recycling from arginine but not for the stress-induced proline accumulation.9 Wilδ-type Arabidopsis plants could utilize either arginine or ornithine as the sole nitrogen source. In oat-knockout mutants, an accumulation of urea cycle intermediates occurred under such nutritional conditions. On the contrary, utilization of urea and stress-induced proline accumulation were not affected by the mutation. This provided strong evidence against a shortcut from arginine to proline that bypasses glutamate and cytosolic P5CS activity. OAT probably links the degradation pathways for arginine and proline (Fig. 2) converging in the formation of GSA/P5C in mitochondria, which is further metabolized to glutamate by P5CDH. It seems that under normal physiological conditions, ornithine can be converted to proline only via glutamate, while this conversion is not contributing substantially to stress-induced proline accumulation.9
Three-dimensional Structure
No crystal structure of a plant OAT has been solved so far. Neverthless, a homology modeling was recently performed for the enzyme from Vigna aconitifolia17 based on the crystal structure of human OAT10 (PDB accession code 1OAT). With this model, a flexible docking study with ornithine and competitive inhibitors was accomplished; see further.
Human liver OAT was first successfully crystallized as a recombinant protein obtained by expression of the entire gene in E. coli.26 Later on, the crystal structure of the enzyme was determined.10,27,28 The functional unit of the protein consists of a dimer built from two identical subunits. Each monomer contains 12 α-helices and 14 β-strands and can be structurally divided into three domains: a large 249 residue domain (PLP-binding domain), a small C-terminal domain of 95 residues and an N-terminal segment of 42 residues. Inter-subunit contacts occur mainly through the large domain.10,27,28 The dimeric enzyme possess two equal active sites (in each subunit), where the cofactor PLP is covalently bound to Lys292 through a Schiff base forming the so-called internal aldimine. The cofactor is located in the large domain at the subunit interface and interacts non-covalently with residues from both subunits. Such a folding is common among several PLP-dependent enzymes.27,28 The packing of the OAT dimers in the crystal yields a hexameric quaternary structure in which three dimers are arranged to form about one turn of a right-handed superhelix.10
Human OAT was also crystallized in the presence of L-canaline and gabaculine, which represent structural analogues of ornithine (the crystal structures are deposited under the PDB accession codes 2CAN and 1GBN, respectively).27 The compounds mimic the natural substrate well enough to bind at the active site and enter the first steps of the transamination reaction. Unlike L-ornithine, however, they cause irreversible inhibition of the enzyme by producing covalent intermediates with the PLP cofactor.27 OAT does not undergo a significant conformational changes upon binding of the inhibitors except for local sidechain movements. Because L-canaline (α-amino-γ-amino-oxybutyric acid) is virtually identical to ornithine, the structure of the enzyme-inhibitor complex allowed discovery of specific residues which participate in substrate recognition via the α-carboxylate and α-amino group (Arg180 and Tyr55, respectively). In this way, OAT specifically discriminates between α- and δ-amino groups of ornithine—only the latter is able to form the reactive Schiff base with PLP cofactor. The same residues were found to be mutated in inactivated OAT of patients suffering from gyrate atrophy.29 Gabaculine (5-amino-1,3-cyclohexadienyl carboxylic acid) binding is facilitated by aromatic-aromatic interactions with two amino acid residues, Tyr85 and Phe177. It has been proposed that a rearrangement (“switch”) of Glu235 at the active site is important for the second OAT half-reaction, where the amino group abstracted by PLP from ornithine is transferred to 2-oxoglutarate as a terminal acceptor.10,28
Another ligand, which was used for co-crystallization of OAT, is (2S, 5S)-5-fluoromethylornithine (5FMOrn). The crystal structure of human OAT complexed with 5FMOrn (PDB accession code 2OAT) provided further information on the binding of ornithine to OAT.28 The fluoro derivative is known as the only inhibitor exclusively specific for ornithine aminotransferase. It blocks the enzyme by a suicide reaction (“mechanism-based inhibition”) leading to a covalent adduct with the cofactor. In the crystal structure, the cofactor adduct is detached from Lys292, but is nonetheless stabilized in its position by several non-covalent interactions with active site residues. The α-carboxylate and the α-amino group of the bound 5FMOrn are anchored to the enzyme by hydrogen bonding to Arg180 and Tyr55, respectively. Although not covalently bound, the adduct is firmly kept at the active site and cannot be removed by dialysis. The other diastereoisomers of 5-fluoromethylornithine cannot bind properly at the recognition site.28
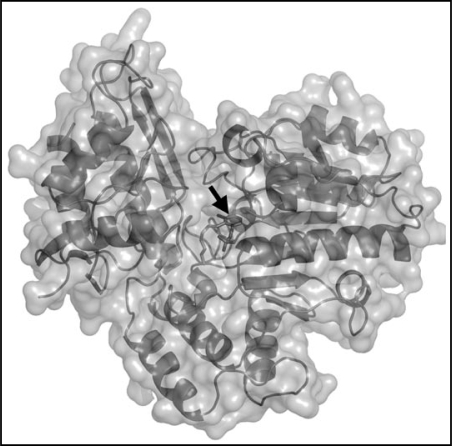
As has been mentioned above, docking studies with the mothbean OAT model revealed interactions that are involved in binding of competitive inhibitors.17 Based on the interaction energies, isoleucine and valine were the best enzyme ligands from this group. Valine showed even more favorable binding energy than ornithine itself. In the case of proline, which displayed no inhibitory properties in experiments performed in vitro, docking calculations revealed the lack of proper binding. Figure 3 shows a three-dimensional structure of pea seedling OAT obtained by homology modeling using human OAT (PDB accession code 1OAT) as a template. There are no significant differences in comparison with the structure of human OAT and the mothbean OAT model.
Figure 3.
A homology-based molecular model of pea seedling OAT. The three-dimensional monomeric structure was modeled with human OAT as a template (PDB accession no. 1OAT) and pea OAT sequence (UniProtKB accession no. B1A0U3) using the program SWISS-MODEL (SIB-Biozentrum Basel, http://swissmodel.expasy.org/). The software used for picture drawing was PyMol (DeLano Scientific LLC, http://pymol.sourceforge.net/). The black arrow aims at the cofactor PLP.
Other Similar ω-Aminotransferases
OAT differs from other aminotransferases for which detailed biochemical information is available (e.g., alanine or aspartate aminotransferases) in that it is specific for an amino group other than the α-amino group adjoining a carboxyl function.10 It belongs to the subgroup III of aminotransferases.30 In the first half reaction, OAT functions as an ω-aminotransferase and attacks the distal δ-amino group of L-ornithine (interestingly the more reactive α-amino group is ignored), while it catalyses an α-transamination in the second half reaction.31 The mechanism by which OAT is able to catalyze these two distinct reactions is not well understood and represents one of the most intriguing aspects of this enzyme. Based on the described crystallographic studies with human OAT10,28 and a mutation study,32 it seems that the reason resides in a rearrangement of the active site. Conversion of the enzyme to its PMP form disrupts the internal Glu235-Arg413 interaction, thus enabling 2-oxoglutarate to be a good substrate in the second half reaction.32
OAT shows structural and mechanistic similarity to other enzymes from the same subgroup like 4-aminobutyric acid aminotransferase (GABA-AT; EC 2.6.1.19) and glutamate-1-semialdehyde aminotransferase (glutamate-1-semialdehyde 2,1-aminomutase; EC 5.4.3.8).27 Interestingly, mutation of Tyr85 in human OAT to Ile, as found in GABA-AT,33 decreased the rate of the reaction of the enzyme with ornithine 1000-fold and increased that with 4-aminobutyrate 16-fold, indicating that Tyr85 is a major determinant of specificity toward ornithine.32 N-acetylornithine aminotransferase from Salmonella typhimurium accepts both N-acetylornithine and ornithine as substrates but has higher affinity to the former one. Structural comparison of the Salmonella enzyme with human OAT suggests that its higher preference for N-acetylornithine may not be due to specific changes in the active site residues but could result from minor conformational changes in some of them.34
There is also a significant similarity of OAT to diamine (or polyamine) aminotransferases, which have been described to act in polyamine metabolism in some bacteria.35–39 It should be noted here that the diamine aminotransferase (DAT; EC 2.6.1.29) from E. coli was described in the middle 60s and that time it was the first known aminotransferase utilizing a substrate with no carboxyl group.35 Figure 4 shows the multiple sequence alignment of several OATs from Leguminosae plants, GABA-AT from A. thaliana and DAT from E. coli. The latter enzyme is able to accept several polyamine substrates including putrescine, cadaverine and spermidine utilizing 2-oxoglutarate, 2-oxobutyrate or pyruvate as terminal amino acceptors.35 The enzyme also converts L-ornithine,36 but it seems to be rather a weak substrate.37 Polyamine aminotransferases were reported in the Gram-positive bacteria Arthrobacter sp. TMP-1,38 and Nocardioides (Arthrobacter) simplex.39 As amino donors, the enzymes accept particularly 1,3-diaminopropane, putrescine, agmatine, spermidine and cadaverine. Pyruvate and 2-oxoglutarate were found to be amino acceptors.
Figure 4.
Amino acid sequence homology of plant OATs. A CLUSTAL W40 multiple sequence alignment is shown comparing plant OATs from pea (Pisum sativum), barrel medic (Medicago truncatula), mothbean (Vigna aconitifolia) and Arabidopsis (Arabidopsis thaliana) with GABA-AT from A. thaliana and DAT from E. coli (annotated as putrescine aminotransferase). The sequences are deposited in the UniProtKB database, the respective accession numbers are given in parentheses. The graphic view was obtained using BioEdit v. 7.0.9.0.41
In microorganisms, the transamination of 1,3-diaminopropane results in 3-aminopropionaldehyde, which is then subjected to aminoaldehyde dehydrogenase (AMADH; EC 1.2.1.19) reaction producing β-alanine.42 3-Aminopropionaldehyde represents the best substrate of plant AMADHs.43 However, still it is unclear, whether the compound is formed predominantly by an oxidative or transamination pathway in plants. The copper-containing diamine oxidase from pea seedlings for example converts 1,3-diaminopropane only negligibly.43 There are some reports on the occurrence of DAT activity in plants,44,45 but to our knowledge, no such an enzyme has been purified and characterized so far. Thus we reasoned that pea OAT, as a presumably similar enzyme, might accept some diamines as substrates. A recombinant pea seedling OAT was obtained by cDNA expression in E. coli and its substrate specificity was measured (Stránská J. et al., unpublished results). The enzyme was found to be strictly specific for L-ornithine showing practically no activity with putrescine, 1,3-diamimopropane and 4-aminobutyrate.
Conclusions
There are two research topics, which elucidate the significance of studying OAT: (i) genetic engineering of plants for increased production of the osmoprotectant proline; (ii) considering human OAT as a potential target for development of new therapeutic drugs. During the last years, interesting data appeared in this context. Future trends will probably involve site-directed mutagenesis of OAT enzymes to better understand the catalytic mechanism and relationships to other members of the same subgroup of aminotransferases.32
Despite the advanced agrotechnologies, which are available these days, drought and salinity are still the major factors affecting crop yield by reducing growth and plant productivity. Genetic engineering allows new ways to develop plants with enhanced osmotolerance and represents an important challenge to improve plant resistance. The accumulation of osmoprotectants has been a target for plant genetic engineering for many years.19 A common strategy that is currently being used in biochemical and experiments is based on overexpressing genes responsible for the biosynthesis of osmoprotectants i.e., coding for enzymes involved in the respective metabolic pathways (choline monooxygenase, betaine aldehyde dehydrogenase, P5CS, etc.,).19,46 It has been shown that transgenic plants overexpressing OAT display enhanced tolerance to salt and drought due to increased proline content.24,47
Human OAT holds a significant scientific interest because of its association with gyrate atrophy, a recessive hereditary genetic dissorder leading to progressive loss of vision and eventually blindness in humans.48 Gyrate atrophy is characterized by elevated ornithine levels arising from OAT dysfunction. Patients suffering from gyrate atrophy usually carry a point mutation in both copies of their OAT gene.29 The OAT mRNA level appears to be normal. There is probably more than one mechanism of OAT inactivation which either involves an incorrect processing of the enzyme or of the correctly processed enzyme.29
Interestingly, human OAT has also been recognized as a potential target for chemotherapeutic drug development. In a study performed to evaluate the effect of selective blocking of mitosis in human cancer cells, OAT was identified as a protein, which binds the antimitotic drug diazonamide A and it has a role in regulating mitotic cell division.49 Diazonamide A induces a G2/M phase growth arrest in a variety of human cancer cell lines, which results from aberrant mitotic spindle formations. It seems that OAT facilitates spindle assembly but the mechanism is unknown. Although the activity may not be essential for normal development, it is critical for cell division in human cancer cells sensitive to diazonamide A.49 The OAT-mediated mechanism of action of diazonamide A in inhibiting cell division implies this compound or their derivatives might show therapeutic activity comparable to clinically used tubulin-binding antimitotics (taxanes and vinca alkaloids) in the absence of toxicity.50
Acknowledgements
This work was supported by the grant no. 522/08/0555 from the Czech Science Foundation.
Abbreviations
- DAT
diamine aminotransferase
- GABA-AT
4-aminobutyric acid aminotransferase
- GSA
L-glutamate γ-semialdehyde
- OAT
ornithine δ-aminotransferase
- P5C
Δ1-pyrroline-5-carboxylate
- PLP
pyridoxal-5′-phosphate
- PMP
pyridoxamine-5′-phosphate
- P5CDH
Δ1-pyrroline-5-carboxylate dehydrogenase
- P5CR
Δ1-pyrroline-5-carboxylate reductase
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
- ProDH
proline dehydrogenase
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6771
References
- 1.Aral B, Kamoun P. The proline biosynthesis in living organisms. Amino Acids. 1997;13:189–217. [Google Scholar]
- 2.Yoshida KM, Juni N, Hori SH. Molecular cloning and characterization of Drosophila ornithine aminotransferase gene. Genes Genet Syst. 1997;72:9–17. doi: 10.1266/ggs.72.9. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda M, Misono H, Soda K, Yonaha K, Toyama S. Purification and crystallization of L-ornithine: α-ketoglutarate δ-aminotransferase from Bacillus sphaericus. FEBS Lett. 1979;105:209–212. doi: 10.1016/0014-5793(79)80613-4. [DOI] [PubMed] [Google Scholar]
- 4.Delauney AJ, Hu CAA, Kavi Kishor PB, Verma DPS. Cloning of ornithine δ-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem. 1993;25:18673–18678. [PubMed] [Google Scholar]
- 5.Strecker HJ. Purification and properties of rat liver ornithine δ-transaminase. J Biol Chem. 1965;240:1225–1230. [PubMed] [Google Scholar]
- 6.Gafan C, Wilson J, Berger LC, Berger BJ. Characterization of the ornithine aminotransferase from Plasmodium falciparum. Mol Biochem Parasitol. 2001;118:1–10. doi: 10.1016/s0166-6851(01)00357-7. [DOI] [PubMed] [Google Scholar]
- 7.Inana G, Totsuka S, Redmond M, Dougherty T, Nagle J, Shiono T, Ohura T, Kominami E, Katunuma N. Molecular cloning of human ornithine aminotransferase mRNA. Proc Natl Acad Sci USA. 1986;83:1203–1207. doi: 10.1073/pnas.83.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roosens NHCJ, Thu TT, Iskandar HM, Jacobs M. Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 1998;117:263–271. doi: 10.1104/pp.117.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funck D, Stadelhofer B, Koch W. Ornithine-δ-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol. 2008:8–40. doi: 10.1186/1471-2229-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen BW, Hennig M, Hohenester E, Jansonius JN, Schirmer T. Crystal structure of human recombinant ornithine aminotransferase. J Mol Biol. 1998;277:81–102. doi: 10.1006/jmbi.1997.1583. [DOI] [PubMed] [Google Scholar]
- 11.Scher WI, Vogel HJ. Occurrence of the ornithine δ-transaminase: a dichotomy. Proc Natl Acad Sci USA. 1957;43:796–803. doi: 10.1073/pnas.43.9.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bone DH. Metabolism of citrulline and ornithine in mung bean mitochondria. Plant Physiol. 1959;34:171–175. doi: 10.1104/pp.34.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazelis M, Fowden L. Conversion of ornithine into proline by enzymes from germinating peanut cotyledons. Phytochemistry. 1969;8:801–809. [Google Scholar]
- 14.Splittstoesser WE, Fowden L. Ornithine transaminase from Cucurbita maxima cotyledons. Phytochemistry. 1973;12:785–790. [Google Scholar]
- 15.Lu TS, Mazelis M. L-ornithine:2-oxoacid aminotransferase from squash (Cucurbita pepo, L.) cotyledons. Purification and properties. Plant Physiol. 1975;55:502–506. doi: 10.1104/pp.55.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Ruiter H, Kollöffel C. Arginine catabolism in the cotyledons of developing and germinating pea seeds. Plant Physiol. 1983;73:525–528. doi: 10.1104/pp.73.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekhar PN, Amrutha RN, Sangam S, Verma DPS, Kavi Kishor PB. Biochemical characterization, homology modeling and docking studies of ornithine δ-aminotransferase—an important enzyme in proline biosynthesis of plants. J Mol Graph Model. 2007;26:709–719. doi: 10.1016/j.jmgm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Bouchereau A, Aziz A, Larher F, Martin-Tanguy J. Polyamines and environmental challenges: recent development. Plant Sci. 1999;140:103–125. [Google Scholar]
- 19.Nuccio ML, Rhodes D, McNeil SD, Hanson AD. Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol. 1999;2:128–134. doi: 10.1016/s1369-5266(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 20.Deuschle K, Funck D, Hellmann H, Däschner K, Binder S, Frommer WB. A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001;27:345–355. doi: 10.1046/j.1365-313x.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- 21.Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaf E, Kunze R, Frommer WB. The role of .1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavi Kishor PB, Sangam S, Amrutha RN, Sri Laxmi P, Naidu KR, Rao KRSS, Rao S, Reddy KJ, Theriappan P, Sreenivasulu N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci. 2005;88:424–438. [Google Scholar]
- 23.Hervieu F, Le Dily F, Huault C, Billard JP. Contribution of ornithine aminotransferase to proline accumulation in NaCI-treated radish cotyledons. Plant Cell Environ. 1995;18:205–210. [Google Scholar]
- 24.Roosens NH, Al Bitar F, Loenders K, Angenon G, Jacobs M. Overexpression of ornithine-δ-aminotransferase increases proline biosynthesis and confers osmotolerance in transgenic plants. Mol Breed. 2002;9:73–80. [Google Scholar]
- 25.Yang CW, Kao CH. Importance of ornithine-δ-aminotransferase to proline accumulation caused by water stress in detached rice leaves. Plant Growth Regul. 1999;27:189–192. [Google Scholar]
- 26.Shen BW, Ramesh V, Mueller R, Hohenester E, Hennig M, Jansonius JN. Crystallization and preliminary X-ray diffraction studies of recombinant human ornithine aminotransferase. J Mol Biol. 1994;243:128–130. doi: 10.1006/jmbi.1994.1637. [DOI] [PubMed] [Google Scholar]
- 27.Shah SA, Shen BW, Brünger AT. Human ornithine aminotransferase complexed with L-canaline and gabaculine: structural basis for substrate recognition. Structure. 1997;5:1067–1075. doi: 10.1016/s0969-2126(97)00258-x. [DOI] [PubMed] [Google Scholar]
- 28.Storici P, Capitani G, Müller R, Schirmer T, Jansonius JN. Crystal structure of human ornithine aminotransferase complexed with the highly specific and potent inhibitor 5-fluoromethylornithine. J Mol Biol. 1999;285:297–309. doi: 10.1006/jmbi.1998.2289. [DOI] [PubMed] [Google Scholar]
- 29.Brody LC, Mitchell GA, Obie C, Michaud J, Steel G, Fontaine G, Robert MF, Sipila I, Kaiser-Kupfer M, Valle D. Ornithine δ-aminotransferase mutations in gyrate atrophy, allelic heterogeneity and functional consequences. J Biol Chem. 1992;267:3302–3307. [PubMed] [Google Scholar]
- 30.Hwang BY, Cho BK, Yun H, Koteshwar K, Kim BG. Revisit of aminotransferase in the genomic era and its application to biocatalysis. J Mol Catal B Enzym. 2005;37:47–55. [Google Scholar]
- 31.Williams JA, Bridge G, Fowler LJ, John RA. The reaction of ornithine aminotransferase with ornithine. Biochem J. 1982;201:221–225. doi: 10.1042/bj2010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markova M, Peneff C, Hewlins MJE, Schirmer T, John RA. Determinants of substrate specificity in ω-aminotransferases. J Biol Chem. 2005;280:36409–36416. doi: 10.1074/jbc.M506977200. [DOI] [PubMed] [Google Scholar]
- 33.Storici P, Capitani G, De Biase D, Moser M, John RA, Jansonius JN, Schirmer T. Crystal structure of GABA-aminotransferase, a target for antiepileptic drug therapy. Biochemistry. 1999;38:8628–8634. doi: 10.1021/bi990478j. [DOI] [PubMed] [Google Scholar]
- 34.Rajaram V, Ratna Prasuna P, Savithri HS, Murthy MRN. Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding. Proteins. 2008;70:429–441. doi: 10.1002/prot.21567. [DOI] [PubMed] [Google Scholar]
- 35.Kim KH. Purification and properties of a diamine α-ketoglutarate transaminase from Escherichia coli. J Biol Chem. 1964;239:783–786. [PubMed] [Google Scholar]
- 36.Yamada H, Kimura T, Tanaka A, Ogata K. Amine transaminase Part I. Properties of amine transaminase of Escherichia coli. Agric Biol Chem. 1964;28:443–450. [Google Scholar]
- 37.Samsonova NN, Smirnov SV, Altman IB, Ptitsyn LR. Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 2003;3:2. doi: 10.1186/1471-2180-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yorifuji T, Kondo S, Shimizu E, Naka T, Ishihara T. Purification and characterization of polyamine aminotransferase of Arthrobacter sp. TMP-1. J Biochem. 1997;122:537–543. doi: 10.1093/oxfordjournals.jbchem.a021786. [DOI] [PubMed] [Google Scholar]
- 39.Kaneoke M, Shimizu E, Yorifuji T. Metabolism of L-arginine, agmatine, and related compounds in Nocardioides simplex. Biosci Biotechnol Biochem. 1994;58:244–249. [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 42.Large PJ. Enzymes and pathways of polyamine breakdown in microorganisms. FEMS Microbiol Rev. 1992;88:249–262. doi: 10.1111/j.1574-6968.1992.tb04991.x. [DOI] [PubMed] [Google Scholar]
- 43.Tylichová M, Kopečný D, Snégaroff J, Šebela M. Aminoaldehyde dehydrogenases: has the time now come for new interesting discoveries? Curr Topics Plant Biol. 2007;8:45–70. [Google Scholar]
- 44.Hasse K, Schmid G. Synthese und Abbau biogener Amine durch enzymatische Transaminierung. Biochem Z. 1963;337:69–79. (Ger). [Google Scholar]
- 45.Michaels R, Kim KH. Comparative studies of putrescine degradation by microorganisms. Biochim Biophys Acta. 1966;115:59–64. doi: 10.1016/0304-4165(66)90048-1. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 2004;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu LQ, Fan ZM, Guo L, Li YQ, Zhang WJ, Qu LJ, Chen ZL. Overexpression of an Arabidopsis δ-OAT gene enhances salt and drought tolerance in transgenic rice. Chin Sci Bull. 2003;48:2050–2056. [Google Scholar]
- 48.Seiler N. Ornithine aminotransferase, a potential target for the treatment of hyperammonemias. Curr Drug Targets. 2000;1:119–153. doi: 10.2174/1389450003349254. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Shang L, Burgett AWG, Harran PG, Wang X. Diazonamide toxins reveal an unexpected function for ornithine δ-amino transferase in mitotic cell division. Proc Natl Acad Sci USA. 2007;104:2068–2073. doi: 10.1073/pnas.0610832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams NS, Burgett AWG, Atkins AS, Wang X, Harran P, McKnight SL. Therapeutic anticancer efficacy of a synthetic diazonamide analog in the absence of overt toxicity. Proc Natl Acad Sci USA. 2007;104:2074–2079. doi: 10.1073/pnas.0611340104. [DOI] [PMC free article] [PubMed] [Google Scholar]