Abstract
An elicitor derived from the cell wall of rice blast fungus (Magnaporthe grisea) causes cell death in suspension cultured cells of rice (Oryza sativa L.). To elucidate the role of M. grisea elicitor on metabolic pathway of rice cells, we performed metabolite profiling using capillary electrophoresis-mass spectrometry (CE/MS). Treatment with M. grisea elicitor increased the amounts of antioxidants and free amino acids and decreased the amount of metabolites in the tricarboxylic acid (TCA) cycle. Lower ATP concentration caused aberrant energy charge, concurrently with reduced amount of NAD(P)H in elicitor treated cells. Among free amino acids detected in this study, the level of gamma-aminobutyric acid (GABA) increased. GABA is metabolized through a bypass pathway of the TCA cycle called GABA shunt, which is composed of glutamate decarboxylase (GAD), GABA transaminase (GABA-T) and succinic semialdehyde dehydrogenase (SSADH). While M. grisea elicitor negligibly affected GAD and SSADH, GABA-T activity significantly decreased. The decrease in GABA-T activity was recovered by NADPH oxidase inhibitor, which prevents cell death induced by M. grisea elicitor. Thus, GABA accumulation observed in rice cells under elicitor stress is partly associated with GABA-T activity.
Key words: metabolome, Magnaporthe grisea, capillary electrophoresis, mass spectrometry, gamma-aminobutyric acid, GABA transaminase, Oryza sativa
Introduction
Plants exert sophisticated defense mechanisms against attacks by pathogens. The defense mechanism is induced when pathogens possessing an avirulent gene attack host plants carrying the resistance gene. These interactions initiate programmed cell death.1–3 Well-described phenomena such as the hypersensitive response (HR) with generation of reactive oxygen species (ROS), nitric oxide and salicylate, can be observed.4–6 Proliferation of pathogens can be prevented by the endogenous synthesis of phytoalexins, pathogenesis-related (PR) proteins, cell wall components, phenol and antioxidants.7–12 Elicitors such as oligosaccharides derived from pathogens induce plant cell death as well as pathogen infection.13 Cell wall extracts derived from Magnaporthe grisea elicit hydrogen peroxide (H2O2) production and cell death in cultured rice cells14 and the upregulation of several defense genes has been reported.15 Using the SAGE method, Matsumura et al.,16 demonstrated transcriptional changes of more than 10,000 genes in rice cells treated with M. grisea elicitor. It is noteworthy that superoxide dismutase (SOD), glutathione-S-transferase (GST) and chitinase were transcriptionally enhanced. On the other hand, OsBI-1, a homolog of Bax inhibitor, was decreased at the mRNA level.17,18 Although elicitors affect the expression of several genes, systematic metabolic profiling has not been reported. Recently, analytical methods such as liquid chromatography-mass spectrometry, gas chromatography-mass spectrometry, nuclear magnetic resonance and capillary electrophoresis-mass spectrometry (CE/MS) provided new insights into the biological function of plant metabolism.19–22 Here, using capillary electrophoresis-mass spectrometry (CE/MS), we determined the concentrations of target metabolites in rice suspension cells treated with M. grisea elicitor to facilitate understanding in the early steps of elicitor-induced processes. Overall changes in the metabolite levels in the central pathways were observed. In addition, M. grisea elicitor affected activities of enzymes in GABA shunt. These results are discussed within the context of relationships between alterations of metabolites and cell death.
Results
Non-target metabolite analysis of rice cultured cells treated with M. grisea elicitor.
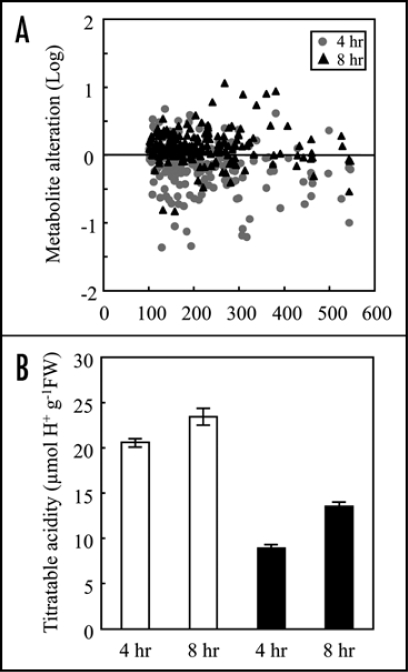
Simultaneous detection using CE/MS is an effective approach for the evaluation of metabolic profiles in rice plants and cultured cells.22,23 We first performed a non-target analysis of metabolites in rice cultured cells treated with elicitor derived from M. grisea (race 007). A mass-to-charge ratio (m/z) range of 100 to 500 was scanned to compare changes in the signal abundances of metabolites after the elicitor treatment (Fig. 1A). The majority of metabolite signals at m/z 100 to 600 were slightly decreased after 4 hrs, while these signals were increased after 8 hrs of elicitor treatment. Among detectable peaks, the ion peaks attributable to GSH (m/z 308) and amino acids such as Ser (m/z 106), Asp (m/z 134) and Gln (m/z 147) increased.
Figure 1.
Metabolic alterations in rice cultured cells induced by M. grisea elicitor. (A) Distribution of intracellular cationic metabolites in rice cultured cells treated with M. grisea elicitor. Metabolite signals were detected by CE/MS at m/z 100 to 600 in a positive ion mode. Relative differences in signal abundances between buffer- or elicitor-treated cells are expressed as the ratios on a logarithmic scale (log10-ratio). Positive and negative log10-ratio values indicate that the abundances were increased or decreased in elicitor-treated cells, respectively. The value close to zero indicates that the abundances were expressed at similar levels. (B) Effect of elicitor on titratable acidity in rice cells. White bars represent cells treated with phosphate buffer and black bars indicate cells treated with M. grisea elicitor. Titratable acidity was measured on homogenized of suspension cultured cells collected at 4 and 8 hrs after the treatments. Titration was performed by 0.01 N NaOH in the presence of phenolphthalein. Data are means ± SD from triplicated experiments.
Titratable acidity, i.e., the contents of titratable organic acid such as citrate and malate,25 was also compared (Fig. 1B). The acidities (µmol H+ g−1FW) in the control cells were 20.57 and 23.46 at 4 and 8 hrs, respectively. After the elicitor treatment, the acidities decreased to 8.90 and 13.55 at 4 and 8 hrs, respectively. These results indicated that titratable organic acid levels in the cells were decreased by the elicitor treatment.
M. grisea elicitor affects the central pathway metabolites in rice cells.
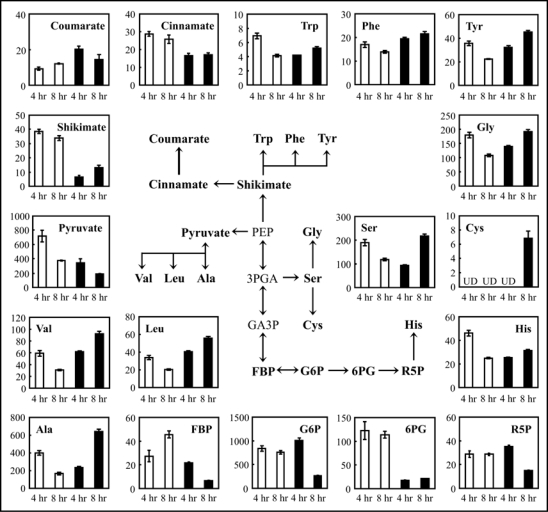
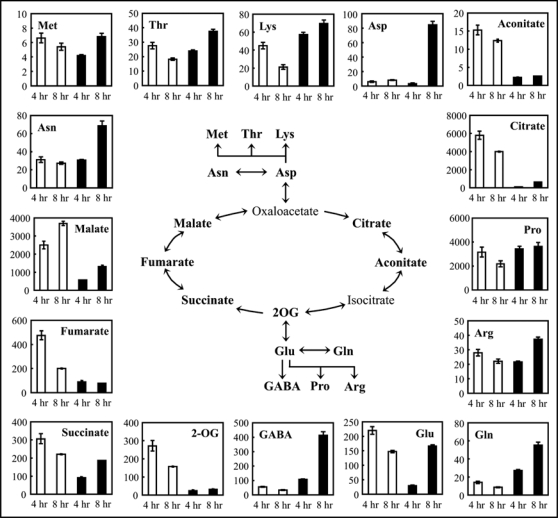
As shown in Figure 1, the results from titratable acidity and non-target analysis suggest that elicitor affected intracellular level of organic acids and amino acids. In order to investigate the early steps of elicitor-induced processes, we quantified primary metabolites that belong to glycolysis, the pentose phosphate (PP) pathway, the shikimate pathway (Fig. 2) as well as the TCA cycle (Fig. 3). Among metabolites in glycolysis, G6P and FBP were unchanged in elicitor-treated suspension cells 4 hrs after treatment (Fig. 2). However, concentration of these metabolites decreased after 8 hrs. Pyruvate levels in elicitor-treated cells decreased to half of controls after both 4 and 8 hrs. On the other hand, pyruvate family amino acids (Ala, Leu, Val) increased after 8 hrs. PEP, 3PGA and GA3P could not be quantified. Levels of metabolites in the PP pathway also decreased in cells treated with elicitor. The level of 6PG was less than 20% of the controls at both 4 and 8 hrs, and R5P was decreased to half of controls at 8 hrs. A different pattern of metabolic alteration was observed in the shikimate pathway. Shikimate was 17% and 39% of the control level at 4 and 8 hrs, respectively. On the other hand, the elicitor induced a 2-fold increase in coumarate level in the cells after 4 hrs. Interestingly, decreases in the levels of metabolites belonging to the TCA cycle were observed in elicitor-treated cells (Fig. 3). After 4 hrs, the metabolites decreased to less than 30% of controls, especially citrate and 2-OG, which decreased to 2% and 9% of controls, respectively. After 4 hrs of elicitor treatment, Glu was decreased to 14% of control, whereas levels of Gln and GABA were 2-fold higher in elicitor-treated cells than controls (Fig. 3). On the other hand, all amino acids detected were increased in elicitor-treated cells at 8 hrs (Figs. 2 and 3). Asp, Gln and GABA were significantly increased; the highest increase (12.5-fold) was observed for GABA.
Figure 2.
Changes in levels of metabolites in rice cells treated with elicitor. Levels of metabolites in glycolysis (G6P, FBP and pyruvate), the pentose phosphate pathway (6PG and R5P), and the shikimate pathway (shikimate, cinnamate and coumarate) as well as amino acids derived from these pathways were compared between buffer-treated suspension cells (white bars) and elicitor-treated suspension cells (black bars), respectively. Values on the y-axis indicate amounts of metabolites (nmol g−1 fresh weight), which are means ± SE calculated from three independent experiments. UD, under detectable level. PEP, 3PGA and GA3P could not be quantified in either elicitor-treated or control cells.
Figure 3.
Quantitative comparison of the TCA cycle metabolites and derived amino acids in rice cultured cells. Metabolite amounts in buffer-treated suspension cells (white bars) and elicitor-treated suspension cells (black bars) were determined at 4 and 8 hrs after treatment. Data are means ± SE from three independent experiments. Oxaloacetate and isocitrate were not detected in neither elicitor-treated or control cells.
M. grisea elicitor affects adenine, pyridine and sugar nucleotide status.
As shown in Figures 2 and 3, metabolites in glycolysis, the PP pathway, and the TCA cycle were decreased by elicitor treatment. Since these pathways require pyridine nucleotides and adenine nucleotides as cofactors, we examined the effect of the elicitor on these nucleotide levels (Table 1). In elicitor-treated cells, NAD increased to 213% of controls at 8 hrs, whereas NADP was elevated to 193% of the control level at 4 hrs and then decreased to the control level at 8 hrs. NADH and NADPH levels slightly decreased at 4 hrs, and significantly decreased at 8 hrs. AMP decreased to about 30% of the controls, while ATP increased to 282% of control at 4 hrs after treatment. However, AMP increased, while ATP decreased to about 40% of control level at 8 hrs after treatment.
Table 1.
Quantitative comparison of nucleotide contents in elicitor-treated rice cells
| Content (nmol g−1 FW) | ||||||
| Metabolites | 4 hours after treatment | 8 hours after treatment | ||||
| Elicitor (−) | Elicitor (+) | Ratio (%) | Elicitor (−) | Elicitor (+) | Ratio (%) | |
| Pyridine nucleotides | ||||||
| NAD | 12.7 | 11.4 | (89) | 14.9 | 31.7 | (213) |
| NADH | 1.8 | 1.0 | (52) | 2.1 | ND | - |
| NADP | 1.4 | 2.7 | (193) | 2.2 | 2.1 | (93) |
| NADPH | 3.5 | 2.6 | (73) | 2.1 | ND | - |
| Adenine nucleotides | ||||||
| AMP | 45.7 | 15.5 | (34) | 13.2 | 37.2 | (281) |
| ADP | 53.2 | 63.9 | (120) | 21.9 | 28.8 | (37) |
| ATP | 45.4 | 128.2 | (283) | 43.4 | 16.0 | (37) |
| Guanine nucleotides | ||||||
| GMP | 2.5 | 2.4 | (98) | 3.2 | 2.9 | (91) |
| GDP | 4.7 | 8.6 | (183) | 3.2 | 6.4 | (200) |
| GTP | 13.9 | 22.4 | (161) | 12.2 | 23.1 | (190) |
| Sugar nucleotides | ||||||
| ADP-Glc | 2.8 | 1.6 | (59) | 1.8 | 1.8 | (98) |
| GDP-Glc | 2.1 | 2.3 | (109) | 1.8 | 1.8 | (98) |
| UDP-Glc | 117.1 | 114.8 | (98) | 116.5 | 107.8 | (93) |
Elicitor (−), buffer-treated callus; Elicitor (+), elicitor-treated callus. Ratio indicates percentage of metabolite contents in Elicitor (+) against that in Elicitor (−). Values are means made on three independent experiments and SE are in all cases <20% of values. ND, not detected.
Pathogen and elicitor affect expression levels of enzymes whose activities depend strongly on sugar nucleotides.29,30 After elicitor treatment, ADP-Glc decreased to 59% of controls at 4 hrs, whereas levels of ADP-Glc, GDP-Glc and UDP-Glc were maintained at the level of the controls.
GABA shunt is rapidly suppressed by M. grisea elicitor treatment.
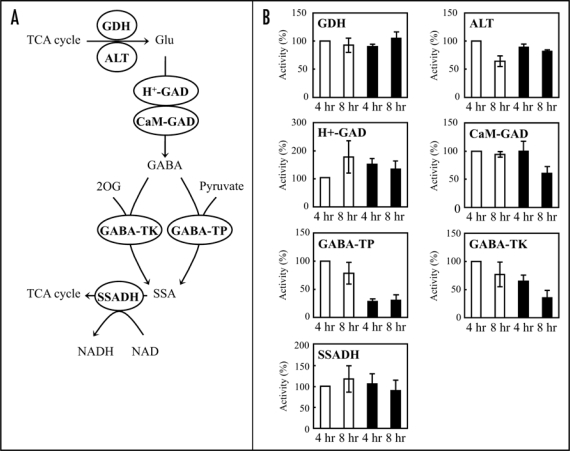
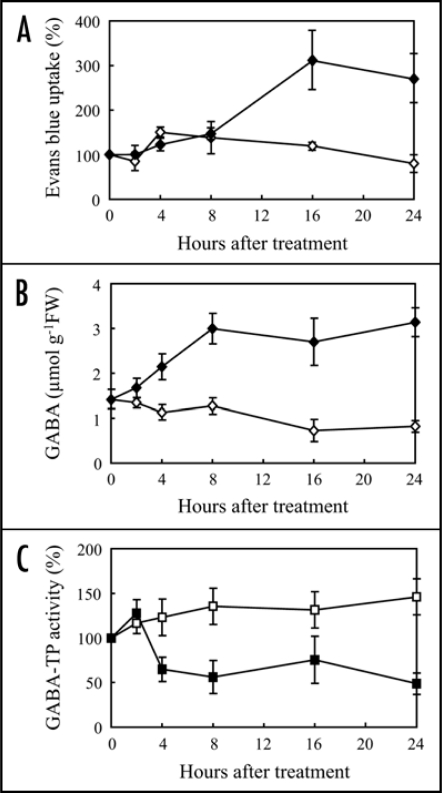
Since elicitor affected TCA cycle metabolites as well as GABA, we focus on the GABA shunt. Following conversion by enzymes such as GDH and ALT, produced Glu is metabolized through the GABA shunt, composed of CaM-GAD, H+-GAD, GABA-TP, GABA-TK, and SSADH (Fig. 4A). After elicitor treatment, activities of GDH and ALT were unchanged, while both CaM-GAD and H+-GAD activities decreased at 8 hrs (Fig. 4B). GABA-TP activity decreased to 28% and 39% of controls at 4 and 8 hrs, respectively. GABA-TK also decreased to 65% and 46% of controls at 4 and 8 hrs, respectively. On the other hand, SSADH activity was almost unaffected at 4 hrs and slightly decreased at 8 hrs. Furthermore, M. grisea elicitor (race 003) also induced cell death (Fig. 5A) and GABA accumulation (Fig. 5B) in rice suspension cultured cells (O. sativa L. cv. Nipponbare). GABA-TP activity slightly increased in control cells, however, the activity in elicitor-treated cells decreased to 50% of control at 4 hrs (Fig. 5C), suggesting that the decrease of GABA-T activity by the elicitor is a common response in rice cells.
Figure 4.
(A) GABA shunt metabolic pathway. Glutamate dehydrogenase (GDH) and alanine transaminase (ALT) produces Glu from TCA cycle intermediate, 2OG. The synthesis of GABA from Glu is catalyzed by two enzymes, acidic pH-dependent glutamate decarboxylase (H+-GAD) and calmodulin-dependent glutamate decarboxylase (CaM-GAD). Then, GABA is converted to succinic semialdehyde (SSA) by GABA transaminases using pyruvate (GABA-TP) or 2OG (GABA-TK) as amino acid acceptors. Finally, succinic semialdehyde dehydrogenase (SSADH) converts SSA to succinate and then succinate enters the TCA cycle. (B) Effects of elicitor on enzyme activities in the GABA shunt. GDH, ALT, H+-GAD, CaM-GAD, GABA-TP, GABA-TK and SSADH activities were measured at 4 and 8 hrs following buffer (White bars) or elicitor treatment (Black bars). Activities are expressed as proportions (%) of the activity in buffer-treated cells at 4 hrs. Data are means ± SD from at least three independent experiments.
Figure 5.
Time course of cell death and GABA-modulatory enzyme activity. The cultured cells from rice (O. sativa L. cv. Nipponbare) were treated with M. grisea elicitor (race 003). (A) Evans blue uptake, (B) GABA accumulation and (C) GABA-TP activity were compared between rice cultured cells treated with phosphate buffer (open symbols) and elicitor (filled symbols). Values in (A and C) are means ± SD from three independent experiments. Values in (B) are means ± SD from three independent experiments. Enzyme activities are expressed as shown in Figure 4.
ROS induced by M. grisea elicitor is associated with the inactivation of GABA-T activity in rice cells.
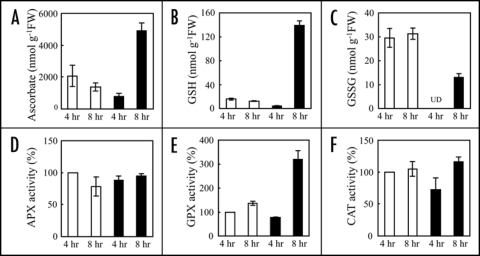
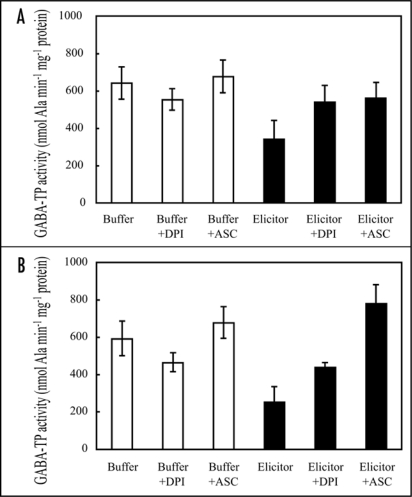
Elicitor causes accumulation of antioxidants in plant cells following ROS production.30–32 In the non-target metabolite analysis, ion peak attributable to GSH was significantly increased. We therefore quantified antioxidant amounts in elicitor-treated rice cells. After elicitor treatment, ascorbate and GSH amounts increased to 357% and 1095% of controls, respectively, at 8 hrs (Fig. 6A and B). The GSSG was lower in elicitor-treated cells than in control cells (Fig. 6C). Since alterations in these antioxidant amounts are closely related to ROS-scavenging enzyme activities, we compared the activities of ascorbate peroxidase (APX), glutathione peroxidase (GPX) and catalase (CAT) between elicitor-treated and control cells. GPX activity was transiently decreased at 4 hrs and then increased to 2-fold of control at 8 hrs (Fig. 6E), whilst APX and CAT activities were remained unchanged (Fig. 6D and F). In order to confirm the relationships between ROS and the elicitor-induced GABA-T inactivation, the effects of an inhibitor of NADPH oxidase and ROS scavengers, on GABA-TP were investigated (Fig. 7). Both 4 and 8 hrs after elicitor treatment, NADPH oxidase inhibitor diphenyleneiodonium (DPI) completely suppressed GABA-TP inactivation (Fig. 7A and B). The elicitor-induced inactivation was also recovered by ascorbate. The treatment of DPI or ascorbate alone had no effect on GABA-Ts activity.
Figure 6.
Effects of elicitor on antioxidant levels and H2O2-scavenging enzyme activities. At 4 and 8 hrs after elicitor-treatment, antioxidants (A–C) and enzyme activities (D–F) were measured. Amounts of ascorbate, GSH and GSSG are represented as shown in Figure 1. APX, GPX and CAT activities were measured using H2O2 as substrate. Changes of activities are expressed as shown in Figure 4. The values are means ± SD calculated from three independent experiments. UD, under detectable level.
Figure 7.
Effect of NADPH oxidase inhibitor and ROS scavenger on GABA-T activity. Rice culture cells were treated with phosphate buffer or elicitor, in the absence or presence of 25 µM diphenyleneiodonium (DPI) or 5 mM ascorbate (ASC). GABA-TP activity was measured at 4 hrs (A) and 8 hrs (B) after the treatment. DPI and ASC were added an hour prier to the buffer and elicitor treatment. Data are means ± SD from three independent experiments.
Discussion
M. grisea elicitor induces drastic alteration of the central pathway metabolites.
We analyzed metabolites using rice suspension cultured cells treated with M. grisea elicitor. In elicitor-treated rice cells, 70% of the detectable metabolites decreased 4 hrs after the treatment. Metabolites in the TCA cycle decreased dramatically, relative to metabolites in other pathways. M. grisea elicitor induces H2O2 production and the expression of ROS-scavenger genes such as SOD and GST;16 antioxidants and ROS-scavenging enzyme activities also increased in our system, suggesting that ROS is involved to the decrease of metabolite levels. Previous study reported that two key mitochondrial enzymes, aconitase and alpha-ketoglutarate dehydrogenase, were the major targets of ROS in mammalian and yeast.33,34 Likewise, ROS induce the degradation of mitochondrial enzymes such as ATP synthase, complex I, succinyl-CoA ligase, aconitase and pyruvate and 2-OG dehydrogenase complexes in plant.35 Therefore, it is possible that the TCA cycle was inhibited and metabolites of the TCA cycle converted to other metabolites or were released to the external environment. Similarly, the amounts of metabolites engaged in glycolysis and the PP pathway decreased after 8 hrs of the elicitor treatment. The decrease in glycolytic metabolites was in agreement with results from SAGE demonstrating the decreased expression of glycolytic enzymes such as phosphoglycerate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase and enolase.16 Therefore, elicitor likely causes the inactivation of the glycolysis pathway through transcriptional and post-translational processes.
The TCA cycle, glycolysis and the PP pathway produce ATP and NAD(P)H, which in turn regulate more than a hundred biochemical reactions in living cells. Elicitor affected nucleotide levels as well as metabolite levels in these pathways. In the early stage of elicitor treatment (4 hrs), energy charge36 was recorded to be 0.772, relative to 0.498 in the control. Thus, under elicitor stress, rice cells may require more ATP through the mitochondrial respiratory machinery. ATP is required not only for the maintenance of cellular functions but also for the activation of protease, which is an essential step in the programmed cell death.37 ATP depletion at a later stage of elicitor treatment (8 hrs) was consistent with previous reports demonstrating that ROS and anoxic stresses caused decreases in ATP level and energy charge.37,38 In addition, SAGE analysis in rice cells treated with elicitor showed that mRNA levels of mitochondrial F1-ATP synthase and nucleoside diphosphate kinase, both ATP-producing enzymes, decreased significantly (data not shown), suggesting that arrest of ATP synthesis was occurred. Furthermore, NADH(P)H concentrations decreased in elicitor-treated cells at 8 hrs. These nucleotides act not only as cofactors of NAD(P)H-requiring pathways such as the respiratory chain, but also as the regulators of cellular redox state. These results may indicate that the cell death is accelerated by ATP depletion and altered redox states in cells.
M. grisea elicitor induces GABA accumulation accompanied by rapid inactivation of GABA-T.
Among free-amino acids, GABA showed highest increasing rate after elicitor treatment. In animals, GABA acts as a neurotransmitter, which has an inhibitory effect on neurons. On the other hand, the function of GABA is unclear in plants. Accumulation of GABA in response to many stresses such as extreme temperature, salt, hypoxia and wounding have been demonstrated.39–43 Therefore, the accumulation of GABA seems to be involved in protection against environmental stresses. GABA is metabolized in a bypass pathway of the TCA cycle called the GABA shunt. The transgenic tomato plants exhibiting decreased activity of succinyl-coenzyme A ligase, the TCA cycle enzyme producing succinate as the respiratory substrate, showed negligible effects on respiration and growth.44 They also noted that flux through the GABA shunt is upregulated in the transgenic plants. Therefore, plants provide a functional bypass to maintain their respiration under a wide range of environmental conditions. The GABA shunt is composed of GAD, GABA-T and SSADH, and converts Glu to succinate.39,45,46 In order to produce Glu, GDH and ALT reductively aminate TCA cycle intermediate, 2-OG, and then Glu is used as a substrate of GAD. The GDH and ALT, which catalyze the reaction between 2-OG and Glu, were not affected by elicitor treatment. This result indicates that the increase in GABA level is not caused by the conversion of metabolites from the TCA cycle to GABA shunt. In the first step of the GABA shunt, two enzymes synthesize GABA from Glu: H+-GAD, activated under acidic conditions (pH < 5.5) and CaM-GAD, a calcium- and CaM-dependent enzyme.42,47 In elicitor-treated cells, these enzymatic activities decreased at 8 hrs compared to the controls. On the other hand, GABA-Ts, GABA-degrading enzymes, were immediately inactivated by the elicitor. The activity of SSADH, the last step of the shunt, did not show a significant change after elicitor treatment. Time course measurement also showed that the increase in GABA level is caused by the prevention of GABA breakdown (Fig. 5). Additional possible cause was the change of permeability of mitochondrial membrane, which in turn release Ca2+ to the cytosol.48,49 Thus, CaM-GAD might be upregulated. Unexpectedly, Ala accumulation was observed in elicitor-treated cells at 8 hrs, although Ala is a byproduct of GABA-T. Miyashita and Good48 reported that Arabidopsis mutants defective in GABA-T activity accumulated Ala at almost same level as wild-type during 4 hrs after hypoxia stress. Therefore, the observed Ala accumulation may be involved in complex series of reactions. The flux analysis in future work will need to reveal this discrepancy. Coleman et al.,49 reported that yeast deficient in the GABA-T gene is highly susceptible to H2O2. Matsumura et al.,16 reported that a NADPH oxidase inhibitor suppressed ROS generation and cell death induced by elicitor. The inactivation of GABA-TP was also suppressed by the inhibitor, indicating that the rapid decrease of GABA-TP activity is associated with ROS-mediated cell death. Furthermore, a high concentration of ascorbate suppressed GABA-TP inactivation, whereas elicitor induced the accumulation of the antioxidant and the activation of ROS scavenging enzymes. The treatment with both elicitor and ascorbate induced GABA-TP activation at 8 hrs. These results suggest that GABA-TP activity can be recovered when the ROS scavenging system is activated. We hypothesize that above a certain threshold, the decoy of the GABA shunt and cell death take place. Our future work will focus on the mechanism regulating relationships between GABA shunt and cell death.
Materials and Methods
Materials.
Rice (Oryza sativa L. cv. Kakehashi) suspension cells were cultured at 28°C in liquid AA medium. Rice (O. sativa L. cv. Nipponbare) suspension cells were cultured according to Takahashi et al.23 Elicitor was extracted from the cell wall of rice blast fungus (M. grisea races 007 and 003) and added to the medium at a final concentration of 1 mg/ml. Cultured cells treated with 20 mM phosphate buffer were used as the control.
CE/MS.
Preparation of samples and determination of metabolites were provided previously in Takahashi et al.24 Briefly, samples were powdered in liquid nitrogen and added to 50% (v/v) ice-cold methanol. After the centrifugation at 15,000 x g for 5 min, the supernatant was filtered through a Millipore 5 kDa cutoff filter (Amicon) and the filtrate was used for analysis. Separation and determination of metabolites were performed with a CE/MS system (Agilent Technologies, Waldbronn, Germany). For the determination of anionic compounds, a polyethylene glycol coated capillary (DB-WAX, J&W Scientific, Folsom, CA) with 20 mM ammonium acetate (pH 6.8) as running buffer was used. Cationic compounds were separated in an uncoated fused-silica capillary using 1 M formic acid (pH 1.9) as running buffer. Anionic and cationic compounds were analyzed in negative ion mode and positive ion mode, respectively. Accuracy was determined by measurement of known concentrations of target compounds.
Titratable acidity.
Acidity of cells was measured by titrating against 0.01 N NaOH with phenolphthalein as indicator.25 Approximately 100 mg of samples was powdered in liquid nitrogen, then 1 ml of 50% methanol preheated to 80°C was added. After centrifugation at 15,000 x g for 5 min, supernatants were used for titration.
Enzymatic assay.
CAT, APX and GPX were assayed by a method of Gabara et al.,26 with minor modifications. Frozen samples were powdered in liquid nitrogen and then extraction buffer (pH 7.0) containing 50 mM potassium phosphate, 1 mM DTT, 10 mM sodium ascorbate and 10% sucrose was added. After centrifugation at 15,000 x g for 5 min, the supernatant was used as an enzyme extract. The CAT activity was quantified by the loss of H2O2 measured at 240 nm. The reaction was started by the addition of 500 mM H2O2 to aqueous solution containing 50 mM potassium phosphate (pH 7.0) and enzyme extract. The APX activity was measured by monitoring the decrease in absorbance at 290 nm. The reaction mixture was composed of 50 mM potassium phosphate, pH 7.0, 5 µM ascorbate and enzyme extract and the reaction was started by the addition of 10 mM H2O2. The GPX activity was measured by following the rate of NADPH oxidation at 340 nm. The reaction mixture contained 50 mM potassium phosphate, 1 mM EDTA, 0.2 mM NADPH, 1 mM GSH, 0.1 mM H2O2, 2 units glutathione reductase and enzyme extract at pH 7.0.
For the measurement of activities of glutamate dehydrogenase (GDH), alanine transaminase (ALT), glutamate decarboxylase (GAD) and gamma-aminobutyrate (GABA) transaminase (GABAT), frozen samples were homogenized in 100 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM DTT and 1 mM EDTA. The homogenate was centrifuged at 15,000 x g for 5 min to obtain supernatant for further analysis. The GDH activity was assayed by incubating enzyme extract in a reaction mixture (pH 7.9) containing 100 mM imidazole, 200 mM ammonium acetate, 1 mM EDTA, 2 mM ADP and 0.2 mM NADH. The reaction was started by the addition of 12 mM 2-oxoglutarate (2OG) and the oxidation was observed at 340 nm. The ALT activity was assayed by determining NADH oxidation in a reaction mixture (pH 7.5) containing 50 mM potassium phosphate, 1 M Ala, 0.1 mM NADH, 10 U lactate dehydrogenase, 2 mM 2OG and enzyme extract. The activities of calmodulin-dependent GAD (CaM-GAD) and acidic pH-dependent GAD (H+-GAD) were measured by determining GABA production. For CaM-GAD activity, the extract was incubated in a reaction mixture (pH 7.5) containing 100 mM HEPES, 1 mM DTT, 1 mM EDTA, 1 mM CaCl2, 50 nM CaM, 5 mM Glu and 0.1 mM pyridoxal-5-phosphate (PLP). For H+-GAD activity, the extract was incubated in a reaction mixture (pH 5.8) containing 100 mM potassium phosphate, 5 mM MgCl2, 1 mM DTT, 5 mM Glu and 0.1 mM PLP. After incubation at 30°C for 15 min, the reaction was quenched by heating at 95°C for 2 min, followed by centrifugation at 15,000 x g for 5 min. The amount of GABA in the resulting supernatant was measured by the CE/MS method described above. Pyruvate-dependent GABA-T (GABA-TP) activity was measured according to the method of Ansari et al.,27 with minor modifications. The extract was added to a reaction mixture (pH 8.2) containing 50 mM Tris-HCl, 1.5 mM DTT, 0.5 mM EDTA, 10% glycerol, 10 mM GABA and 0.1 mM PLP. After addition of 4 mM pyruvate, the solution was incubated at 30°C for 1 hr and the amount of Ala formed was quantified by CE/MS. For the measurement of 2OG-dependent GABA-T (GABA-TK) activity, 4 mM pyruvate was replaced to 4 mM 2OG and produced Glu was quantified. Succinic semialdehyde dehydrogenase (SSADH) activity was measured according to Busch and Fromm28 with minor modifications. Frozen samples were homogenized in 30 mM borate buffer (pH 9.0) containing 1 mM DTT, centrifuged at 15,000 x g for 5 min, and resulting supernatant was used kept for further analysis. The supernatant was incubated in reaction mixture (pH 9.0) containing 30 mM borate, 0.5 mM NAD and 0.1 mM succinic semialdehyde. After incubation at 24°C for 30 min, the reaction was terminated by heating at 95°C for 2 min and centrifugation at 15,000 x g for 5 min. The amount of succinate in the resulting supernatant was measured by the CE/MS.
Protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad Laboratories, CA., USA).
Reference reagents.
Fructose-1,6-bisphosphate (FBP), 3-phosphoglycerate (3PGA), glyceraldehyde-3-phosphate (GA3P), phosphoenolpyruvate (PEP), pyruvate, citrate, aconitate, isocitrate, 2-OG, succinate, fumarate, malate, glucose-6-phosphate (G6P), 6-phosphogluconate (6PG), ribose-5-phosphate (R5P), shikimate, cinnamate, coumarate, ascorbate, reduced glutathione (GSH), oxidized glutathione (GSSG), GABA, nucleotides, nucleotide sugars and amino acids were analytical grade. The 20 µM standard solutions were prepared for each compound.
Acknowledgements
Intensive edition of this manuscript by Dr. M.S. Uchimiya is appreciated. This research was supported in part by a grant from the Ministry of Agriculture, Forestry and Fishery, Japan and CREST, JST, Japan.
Abbreviations
- CE/MS
capillary electrophoresis mass spectrometry
- GABA
gamma-aminobutyric acid
- GABA-T
GABA transaminase
- ROS
reactive oxygen species
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6112
References
- 1.Keen NT. Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 2.Grant M, Mansfield J. Early events in host-pathogen interactions. Curr Opin Plant Biol. 1999;2:312–319. doi: 10.1016/S1369-5266(99)80055-7. [DOI] [PubMed] [Google Scholar]
- 3.Jones DA, Takemoto D. Plant innate immunity—direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol. 2004;16:48–62. doi: 10.1016/j.coi.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Able AJ, Guest DI, Sutherland MW. Relationship between transmembrane ion movements, production of reactive oxygen species and the hypersensitive response during the challenge of tobacco suspension cells by zoospores of Phytophthora nicotianae. Physiol Mol Plant Pathol. 2001;58:189–198. [Google Scholar]
- 5.Río LA, Corpas FJ, Barroso JB. Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry. 2004;65:783–792. doi: 10.1016/j.phytochem.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Smirnov S, Shulaev V, Tumer NE. Expression of pokeweed antiviral protein in transgenic plants induces virus resistance in grafted wild-type plants independently of salicylic acid accumulation and pathogenesis-related protein synthesis. Plant Physiol. 1997;114:1113–1121. doi: 10.1104/pp.114.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissey JP, Osbourn AE. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol Mol Biol Rev. 1999;63:708–724. doi: 10.1128/mmbr.63.3.708-724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benhamou N, Kloepper JW, Quadt Hallman A, Tuzun S. Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol. 1996;112:919–929. doi: 10.1104/pp.112.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Ascensao ARFDC, Dubery IA. Soluble and wall-bound phenolics and phenolics polymers in Musa acuminata roots exposed to elicitors from Fusarium oxysporum f.sp. cubense. Phytochemistry. 2003;63:679–686. doi: 10.1016/s0031-9422(03)00286-3. [DOI] [PubMed] [Google Scholar]
- 11.Pinto MC, Tommasi F, Gara LD. Changes in the antioxidant systems as part of the signaling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco bright-yellow 2 cells. Plant Physiol. 2002;130:698–708. doi: 10.1104/pp.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker CJ, Mock NM. A method to detect oxidative stress by monitoring changes in the extracellular antioxidant capacity in plant suspension cells. Physiol Mol Plant Pathol. 2004;64:255–261. [Google Scholar]
- 13.Hahn MG. Microbial elicitors and their receptors in plants. Annu Rev Phytopathol. 1996;34:387–412. doi: 10.1146/annurev.phyto.34.1.387. [DOI] [PubMed] [Google Scholar]
- 14.Kim ST, Cho KS, Yu S, Kin SG, Hong JC, Han CD, Bae DW, Nam MH, Kang KY. Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics. 2003;3:2368–2378. doi: 10.1002/pmic.200300577. [DOI] [PubMed] [Google Scholar]
- 15.Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA. 2001;98:759–764. doi: 10.1073/pnas.021273498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R. Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J. 2003;33:425–434. doi: 10.1046/j.1365-313x.2003.01639.x. [DOI] [PubMed] [Google Scholar]
- 17.Kawai M, Pan L, Reed JC, Uchimiya H. Evolutionally conserved plant homologue of the Bax Inhibitor-1 (BI-1) gene capable of suppressing Bax-induced cell death in yeast. FEBS lett. 1999;464:143–147. doi: 10.1016/s0014-5793(99)01695-6. [DOI] [PubMed] [Google Scholar]
- 18.Kawai-Yamada M, Ohori Y, Uchimiya H. Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell. 2004;16:21–32. doi: 10.1105/tpc.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 20.Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell. 2000;13:11–29. doi: 10.1105/tpc.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirai MY, Yano M, Goodenowe D, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato S, Soga T, Nishioka T, Tomita M. Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 2004;40:151–163. doi: 10.1111/j.1365-313X.2004.02187.x. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi H, Hayashi M, Goto F, Sato S, Soga S, Nishioka T, Tomita M, Kawai Yamada M, Uchimiya H. Evaluation of metabolic alteration in transgenic rice overexpressing dihydroflavonol-4-reductase. Ann Bot. 2006;98:819–825. doi: 10.1093/aob/mcl162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi H, Watanabe A, Tanaka A, Hashida SN, Kawai Yamada M, Sonoike K, Uchimiya H. Chloroplast NAD kinase is essential for energy transduction through the xanthophyll cycle in photosynthesis. Plant Cell Physiol. 2006;47:1678–1682. doi: 10.1093/pcp/pcl029. [DOI] [PubMed] [Google Scholar]
- 25.Taybi T, Nimmo HG, Borland AM. Expression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase genes. Implications for genotypic capacity and phenotypic plasticity in the expression of crassulacean acid metabolism. Plant Physiol. 2004;135:587–598. doi: 10.1104/pp.103.036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabara B, Sklodowska M, Wyrwicka A, Glinska S, Gapinska M. Changes in the ultrastructure of chloroplasts and mitochondria and antioxidant enzyme activity in Lycopersicon esculentum Mill. Leaves sprayed with acid rain. Plant Sci. 2003;164:507–516. [Google Scholar]
- 27.Ansari MI, Lee RH, Chen SCG. A novel senescence-associated gene encoding γ-aminobutyric acid (GABA):pyruvate transaminase is upregulated during rice leaf senescence. Physiol Plant. 2005;123:1–8. [Google Scholar]
- 28.Busch KB, Fromm H. Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol. 1999;121:589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Idnurm A, Warnecke DC, Heinz E, Howlett BJ. Characterisation of neutral trehalase and UDP-glucose:sterol glucosyltransferase genes from the plant pathogenic fungus Leptosphaeria maculans. Physiol Mol Plant Pathol. 2003;62:305–313. [Google Scholar]
- 30.Chong J, Baltz R, Fritig B, Saindrenan P. An early salicylic acid-, pathogen- and elicitor-inducible tobacco glucosyltransferase: role in compartmentalization of phenolics and H2O2 metabolism. FEBS Lett. 1999;458:204–208. doi: 10.1016/s0014-5793(99)01154-0. [DOI] [PubMed] [Google Scholar]
- 31.Hirasawa K, Amano T, Shioi Y. Effects of scavengers for active oxygen species on cell death by cryptogein. Phytochemistry. 2005;66:463–468. doi: 10.1016/j.phytochem.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Pauw B, van Duijin B, Kijne JW, Memelink J. Activation of the oxidative burst by yeast elicitor in Catharanthus roseus cells occurs independently of the activation of genes involved in alkaloid biosynthesis. Plant Mol Biol. 2004;55:797–805. doi: 10.1007/s11103-004-1968-2. [DOI] [PubMed] [Google Scholar]
- 33.Tretter L, Adam Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci. 2005;360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 35.Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002;32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- 36.Atkinson DE. Cellular energy metabolism and its regulation. New York: Academic Press; 1977. pp. 85–107. [Google Scholar]
- 37.Tiwari BS, Belenghi B, Levine A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002;128:1271–1281. doi: 10.1104/pp.010999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia JH, Saglio P, Roberts JKM. Nucleotide levels do not critically determine survival of maize root tips acclimated to a low-oxygen environment. Plant Physiol. 1995;108:589–595. doi: 10.1104/pp.108.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- 40.Pérez Alfocea F, Santa Cruz A, Guerrier G, Bolarin MC. NaCl stress-induced organic solute changes on leaves and calli of Lycopersicon esculentum, L. pennellii and their interspecific hybrid. J Plant Physiol. 1994;143:106–111. [Google Scholar]
- 41.Bown AW, Shelp BJ. The metabolism and functions of γ-aminobutyric acid. Plant Physiol. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinnersley AM. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci. 2000;19:479–509. [Google Scholar]
- 43.Rentsch D. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studart Guimaraes C, Fait A, Nunes Nesi A, Carrari F, Usadel B, Fernie AR. Reduced expression of succinyl-coenzyme A ligase can be compensated for by upregulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 2007;145:626–639. doi: 10.1104/pp.107.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouché N, Fromm H. GABA in plants: just a metabolite? Trens Plant Sci. 2004;9:110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Fait A, Fromm H, Walter D, Galili G, Fernie AR. Highway or byway: the metabolic role of the GABA shunt in plants. Trens Plant Sci. 2007;13:14–19. doi: 10.1016/j.tplants.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Snedden WA, Koutsia N, Baum G, Fromm H. Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- 48.Duchen MR. Mitochondria and Ca2+ in cell physiology and pathophysiology. Cell Calcium. 2000;28:339–348. doi: 10.1054/ceca.2000.0170. [DOI] [PubMed] [Google Scholar]
- 49.Paolo B, Petronilli V. The permeability transition pore as a mitochondrial calcium release channel: A critical appraisal. J Bioenerg Biomembr. 1996;28:131–138. doi: 10.1007/BF02110643. [DOI] [PubMed] [Google Scholar]
- 50.Miyashita Y, Good AG. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:92–102. doi: 10.1093/pcp/pcm171. [DOI] [PubMed] [Google Scholar]
- 51.Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye Rowley WS. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]