Abstract
In Arabidopsis thaliana, a eudicot species, the transcription factor LFY is expressed throughout the floral meristem and promotes their formation. The expression pattern of the rice LFY homolog-RFL shows distinct differences from that of its Arabidopsis counterpart. In the March issue of PNAS (2008) we have shown the temporally-regulated high-level expression of RFL in the apical meristem is necessary for its transition to an inflorescence meristem and thus to initiate flowering. RFL controls the time taken for flowering, by activating integrators of flowering signals such as OsSOC1 and RFT1. Further, the dynamic pattern of RFL expression in the branching inflorescence meristem (panicle) and in vegetative axillary meristems (tiller buds) is required for panicle branching and tiller outgrowth. Thus RFL functions determine the architecture of the rice plant. Here we propose a plausible model for a regulatory feedback loop between RFL and OsSOC1/RFT1 in controlling the vegetative to flowering phase transition. We discuss the possibility that non-cell autonomous RFL functions may also regulate signaling the net outcome of which determines the rice plant body plan.
Key words: RFL, rice flowering time, plant architecture, signaling, regulatory feedback loop
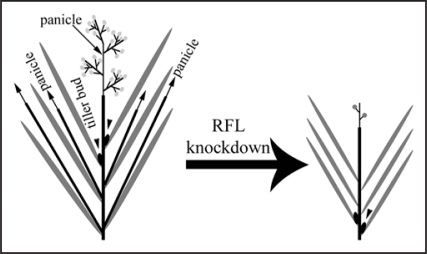
Flowering- a dramatic event in a flowering plants life-cycle is triggered by both environmental cues and intrinsic factors. In Arabidopsis signals from a complex network of activators and repressors are integrated by a key signaling molecule (FT) and a transcription factor (SOC1) to activate high-level LFY expression on the flanks of the shoot apex. This in turn programs a floral meristem by upregulating floral organ patterning genes on the flanks of the apical meristem.1,2 In lfy mutants an inflorescence is made but solitary flowers are replaced by new meristems with characteristics between floral and inflorescence meristems.3 LFY homologs, from many plant species, share some common expression domains for e.g., that in young floral meristems. Yet many homologs display certain distinct profiles. The expression profile of rice RFL is strikingly different from that of any other LFY-like gene. RFL expression in the apical inflorescence meristem, its down regulation at anlagen for branch primordia, expression in outgrowing branches followed by low-level expression in young floret primordia is known for a decade now.4,5 This profile bears some similarity to that of homeodomain-containing meristem function genes.6 Our recent report7 demonstrates the necessity of this dynamic profile for switching the vegetative apical meristem to an inflorescence fate; with its subsequent expression regulating inflorescence branching. Additionally, RFL expression in the axils of leaves and in young axillary meristems, at the basal nodes of a plant, explained the phenotypic effects of reduced tillering seen on its knockdown (Fig. 1).
Figure 1.
Schematic diagram showing the effects of RFL knockdown on rice shoot architecture: black lines, main stem with panicle and tillers with secondary panicles; grey lines, leaves; filled black ovals, tiller buds; solid grey circles, spikelets on the branched panicle. For details, see the text.
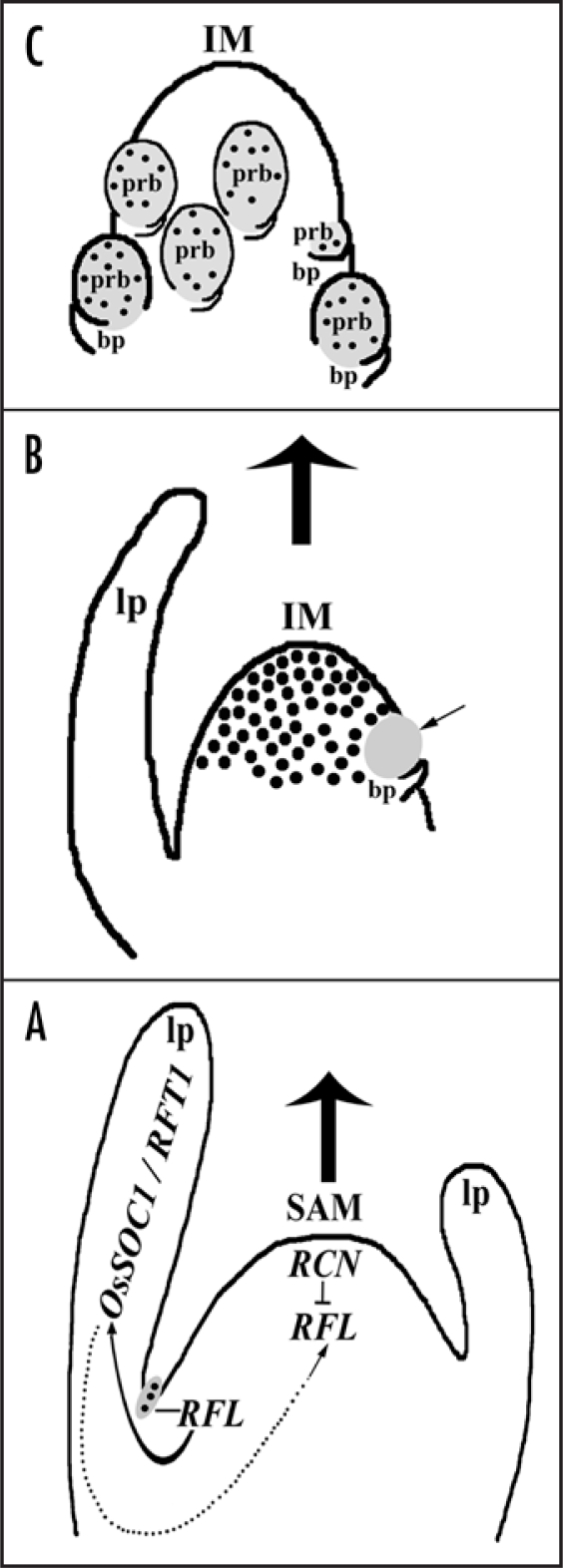
How may flowering regulators be networked to generate the typical rice inflorescence pattern? Our study demonstrates RFL as a regulator of some rice flowering signals. Since transcript levels of OsSOC1 and RFT1 are affected by the RFL expression status we infer these genes act downstream to RFL to trigger phase transition. We speculate a possible feedback loop wherein RFL expression in vegetative axillary meristems activates, in the leaf, the above mentioned flowering signal integrators. This in turn sets up a mechanism to up regulate RFL in the main apical meristem thereby promoting its transition to an inflorescence meristem (Fig. 2). While our study places RFL as a regulator of rice flowering integrators, the contribution of the RFL-mediated pathway in photoperiod-sensitive rice varieties needs examination. This will elucidate if RFL acts parallel to or synergistically with the photoperiod-regulated pathway in activating flowering time integrators. In Arabidopsis the antagonistic interaction between TFL1 and LFY controls the switch from vegetative to reproductive phase.8 A similar scenario can occur in rice, as the TFL1 homologs RCN1 and RCN2 may inhibit RFL expression in vegetative shoot apex and in turn RFL in the inflorescence meristem may exclude RCN1 and RCN2 in these cells in order to initiate flowering (Fig. 2). The similarity in the delayed flowering phenotype seen on overexpression of RCN1 or RCN29 or on the loss of RFL7 lends support to this hypothesis. All together the cascade of temporally and spatially regulated RFL expression in the apical inflorescence meristem may act to initially maintain a meristematic state and with its subsequent repression allowing a transition to another state as predicted by Prusinkiewicz et al.,10 from their modeling of the evolution of inflorescence forms.
Figure 2.
Model predicting the molecular events during vegetative to reproductive phase transition. (A) RFL expression in the axillary meristems (grey oval with dots) activates (arrow with solid line) flowering integrators which can activate RFL in the apical meristem (arrow with dotted line). RFL in the SAM counters flowering repressors as discussed in the text. (B) RFL expression in the newly formed inflorescence meristem (black dots) and its repression at sites of new branch primordia (grey oval indicated by an arrow). (C) RFL expression in outgrowing primary inflorescence branches (grey ovals with dots) formed in the axils of bracts. lp, leaf primordia; bp, bract primordia; IM, inflorescence meristem; prb, primary rachis branch primordia.
Can RFL expression in vegetative axillary meristems have an effect on the shoot apical meristem (SAM) where it is not expressed? One can postulate such a role in the light of studies on the phan mutant of Antirhhinum majus11 and of the Arabidopsis polarity genes FIL and YAB3.12 Though PHAN is expressed in leaf primordia its effects on apical meristem are non-cell autonomous.11 Similarly, manipulation of FIL or YAB3 which induce abaxial cell-fate and or of AGO1 and its closely related protein PINHEAD/ZWILLE which specify adaxial fates in leaf primordium, have effects on the SAM. These data reflect a close relationship between emerging lateral primordia and stem cell function13 and suggest that the leaf primordium may produce a signal that maintains SAM activity. Similarly the direct or indirect effects of RFL on the SAM can be explained by a model in which axillary meristem cells expressing RFL transmit a signal to regulate genes involved in maintenance of the apical meristem.
Polarized gradients of auxin have been recently proposed as a common module for lateral organ formation.14 Our data provide some leads on how RFL knockdown may influence the emergence of lateral primordia by perturbing this gradient. A predicted rice polar auxin efflux facilitator, OsPIN3-like (AK101504), is expressed in domains that overlap with RFL especially in vegetative and reproductive lateral meristems and its transcripts are downregulated upon RFL knockdown. The decreased expression of OsPIN3-like is physiologically reflected in the hypersensitivity of plants with very weak RFL knockdown phenotypes to low concentrations of auxin gradient inhibitors (our unpublished data). This throws up the possibility of the RFL-PIN3 loop as a candidate for a shared mechanism controlling panicle branching and as well as tiller bud outgrowth.
Does RFL coordinate multiple hormonal signals to regulate shoot architecture? A generalized effect of RFL in auxin-signaling cannot be ruled out as in RFL knockdown plants the transcript levels for several cytochrome P450s are affected. Notable among them are some that may affect signaling directly or indirectly. For e.g., transcript levels for predicted rice CYP51G1 gene-family member7,15 is significantly reduced which may cause change in membrane sterol content. Maintaining appropriate membrane sterol compositions is critical for signaling events as underscored by the developmental defects of Arabidopsis mutants for CYP51A2- a member of the same subfamily.16 Not surprisingly, reduced membrane sterols levels disturb the localization of auxin efflux proteins and perturb auxin signaling.17,18 Though none of the RFL-regulated cytochrome P450s genes7 are directly involved in Brassinosteroid (BR) or Gibberellins (GA) biosynthetic pathways, it is possible that levels of precursor molecules that feed into these metabolic pathways may be affected as Arabidopsis homologs for few of these genes are known to act in steroid biosynthesis.17 Also downregulated, in RFL knockdown plants,7 are seven genes of CYP86 subfamily which are annotated as fatty acid hydroxylation factors. These genes are likely to reflect altered cuticle/epidermal wax patterns formed in knockdown panicles. Thus probing in greater depth the mechanism by which RFL coordinates multiple hormone signaling pathways should further our understanding of the cross talk between transcription factors and signaling that regulates development.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6174
References
- 1.Lohmann JU, Weigel D. Building Beauty: The Genetic Control of Floral patterning. Developmental Cell. 2002;2:135–142. doi: 10.1016/s1534-5807(02)00122-3. [DOI] [PubMed] [Google Scholar]
- 2.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 3.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 4.Kyozuka J, Konishi S, Nemato K, Izawa T, Shimamoto K. Downregulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad K, Kushalappa K, Vijayraghavan U. Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence. Mech Dev. 2003;120:491–502. doi: 10.1016/s0925-4773(02)00457-4. [DOI] [PubMed] [Google Scholar]
- 6.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 7.Rao NN, Prasad K, Kumar PR, Vijayraghavan U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA. 2008;105:3646–3651. doi: 10.1073/pnas.0709059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ. A common mechanism controls the life cycle and architecture of plants. Development. 1998;125:1609–1615. doi: 10.1242/dev.125.9.1609. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 10.Prusinkiewicz P, Erasmus Y, Lane B, Hardner LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 11.Waites R, Selvadurai HRN, Oliver IR, Hudson A. The PHANTASTICA Gene Encodes a MYB Transcription Factor Involved in Growth and Dorsoventrality of Lateral Organs in Antirrhinum. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- 12.Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- 13.Kidner CA, Martienssen RA. The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol. 2005;280:504–517. doi: 10.1016/j.ydbio.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 14.Benkova E, Michniewicz M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux depend auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 15.Nelson DR, Schuler MA, Paquette SM, Reichhart DW, Bak S. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. Plant Physiol. 2004;135:756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann FA, Feyereisen R. Arabidopsis cyp51 Mutant Shows Postembryonic Seedling Lethality Associated with Lack of Membrane Integrity. Plant Physiol. 2005;138:2033–2047. doi: 10.1104/pp.105.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willemsen V, Friml J, Grebe M, Van den Toorn A, Palme k, Scheres B. Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell. 2003;15:612–625. doi: 10.1105/tpc.008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souter M, Topping J, Pullen M, Friml J, Hackett R, Grierson D, Lindsey K. hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell. 2002;14:1017–1031. doi: 10.1105/tpc.001248. [DOI] [PMC free article] [PubMed] [Google Scholar]