Abstract
The strategies used by necrotrophic fungal pathogens to infect plants are often perceived as lacking the sophistication of their haustorium producing, host defence suppressing, biotrophic counterparts. There is also a relative paucity of knowledge regarding how effective gene-for-gene based resistance reactions might function against necrotrophic plant pathogens. However, recent data has emerged from a number of systems which has highlighted that particular species of necrotrophic (and/or hemibiotrophic) fungi, have evolved very sophisticated strategies for plant infection which appear, in fact, to hijack the host resistance responses that are commonly deployed against biotrophs. Both disease resistance (R) protein homologues and mitogen-activated protein kinase (MAPK) cascades commonly associated with incompatible disease resistance responses; appear to be targeted by necrotrophic fungi during compatible disease interactions. These findings highlight an emerging sophistication in the strategies deployed by necrotrophic fungi to infect plants.
Key words: Mycosphaerella graminicola, Septoria tritici, Triticum aestivum, mitogen-activated protein kinase, programmed cell death, fungal pathogen, disease resistance, disease susceptibility, toxin
Introduction
Enormous progress has been made within the past two decades in understanding the mechanisms of disease resistance of plants towards pathogens, particularly through studies on “model” systems.1 However, arguably there has been an overall experimental bias towards investigating plant resistance reactions towards biotrophic pathogens, which require living host tissue to complete their infection cycle. Biotrophic fungal and oomycete pathogens have evolved sophisticated methods for acquiring nutrition from living host cells, whilst also suppressing the defence reactions that might otherwise have been triggered in these cells. In contrast to this widely appreciated sophistication, necrotrophic fungal pathogens are often thought of as quite unsophisticated “rotters” with relatively unselective tissue and host specificity. A common opinion regarding how necrotrophic fungal pathogens infect plants is that they simply bludgeon host tissues to death through the combined action of cell wall degrading enzymes and toxins. This concept has recently begun to be challenged though. For example, a disease resistance protein homologue which is targeted by a fungal toxin and plays an important role in facilitating plant infection2 was recently identified in Arabidopsis. This study suggested that necrotrophic host-specific toxin producing fungi might actually hijack elements of the plant defence signalling machinery.
It is generally assumed that plant resistance to biotrophic pathogens is aided by the hypersensitive response (HR) which is a form of programmed cell death (PCD).3,4 HR often occurs as a consequence of the activation of intricate signal transduction pathways that can involve, for example, ERK-like (extracellular-signal regulated kinase) mitogen-activated protein kinases (MAPKs) homologous to Arabidopsis AtMPK6 and AtMPK3. HR-like cell death and MAPK activation has been shown on many occasions to feature in incompatible (disease resistance) interactions triggered following pathogen recognition through a number of both plasma membrane bound and intracellular disease resistance proteins.5–9
Mycosphaerella graminicola—A Sophisticated Necrotrophic Fungal Pathogen
Mycosphaerella graminicola is a wheat leaf-specific, non cell-penetrating, yet ultimately necrotrophic fungal pathogen, and the causal agent of Septoria tritici blotch disease. Specific disease resistance reactions between M. graminicola and wheat have been shown to conform to the gene-for-gene hypothesis.10,11 Somewhat unusually for a necrotrophic plant pathogen, M. graminicola has a relatively long latent period (>8–9 days) before inducing disease symptoms on a susceptible host genotype (compatible interaction).12 The disease symptoms themselves take the form of strictly localised lesions within which the fungus sporulates asexually. Conversely, M. graminicola will often cause no disease symptoms whatsoever (neither macroscopic nor microscopic) on a resistant genotype (incompatible interaction) and it ultimately fails to sporulate. Recent data,12 and the current article, highlight that many of the host responses triggered during a compatible interaction between wheat leaves and M. graminicola are remarkably similar to those implicated in providing resistance to biotrophic plant pathogens. This includes a strictly localised HR-like form of PCD which is preceded by a strong transcriptional activation of the wheat AtMPK3 homologue, TaMPK3 and the subsequent accumulation of the protein. As HR-like symptoms appear during the compatible interaction between wheat and M. graminicola, the TaMPK3 protein is post-translationally activated coincident with the appearance of PCD markers. As previously mentioned, these events have been reported to feature in a number of incompatible interactions between plants and pathogens.5–9 In contrast, during incompatible interactions between wheat and M. graminicola no PCD occurs and there are no significant alterations in TaMPK3 transcript, protein or protein activity levels.
In model dicot systems, AtMPK3 and AtMPK6 homologues are often simultaneously activated via the same MAPK-kinase (MKK).14–16 In some cases these kinases have also been shown to exhibit functional redundancy. However, the wheat homologue of AtMPK6, referred to as TaMPK6, is not activated at any stage of compatible or incompatible interactions with M. graminicola. During a compatible interaction the level of TaMPK6 protein actually begins to decrease around the time that the TaMPK3 protein begins to accumulate. When TaMPK3 subsequently becomes post-translationally activated, TaMPK6 is hardly detectable in cell extracts. It is thus doubtful that TaMPK3 and TaMPK6 will have redundant functions during wheat leaf infection by M. graminicola. This also raises questions as to whether a single MKK will serve to activate both TaMPK6 and TaMPK3 in wheat, or in monocots in general. The sequenced genome of rice contains only five ERK-like MAPKs, which contrasts with the twelve found in Arabidopsis.17 Our own examination of all publicly available wheat EST's in addition to a proprietary collection also identified only five different sequences with homology to different Arabidopsis ERK-like MAPK's. This may suggest that generations of conventional crop breeding has resulted in a smaller functional gene family being retained in modern cereal varieties than perhaps once existed in wild ancestors.
A Model for Wheat Leaf Susceptibility and Resistance Towards M. graminicola
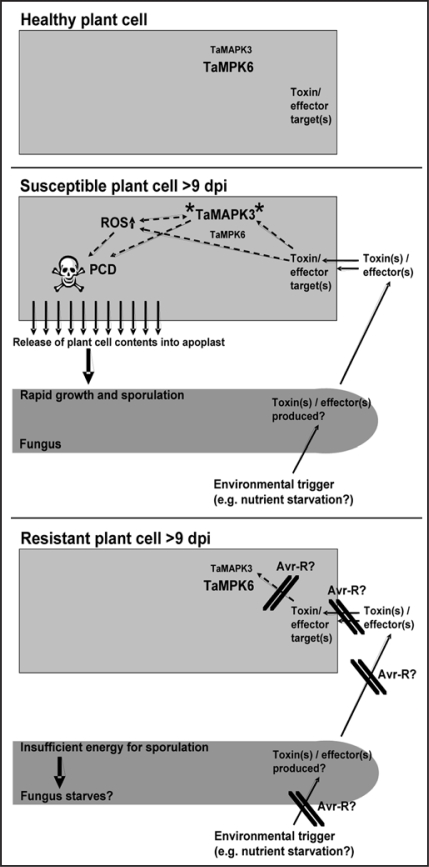
So how does isolate-specific resistance of wheat towards M. graminicola operate? As highlighted in the current paper, much of what we have learnt to date about the mechanism of disease resistance in this system is that there appears to be a distinct lack of “susceptibility signalling” reactions. There is neither localised PCD nor effects imposed upon the TaMPK3 and TaMPK6 protein kinases during an incompatible interaction. Predicting how and where a specific resistance gene product (referred to as Stb genes) functions is further complicated by the fact that even during the symptomless phase (up to ∼9 days post inoculation) of the compatible interaction very little fungal biomass accumulates. Obviously this is also true for the incompatible interactions. Significant increases in fungal growth only occur coincident with PCD and TaMPK3 activation during the compatible interaction. Neither of these events occurs during incompatible interactions and fungal biomass never appreciably increases. Based on these observations we can propose a model for compatible and incompatible interactions in this system (Fig. 1). Compatible interactions result from the fungus hijacking disease resistance signalling pathways (which also perhaps includes an oxidative burst response18) that are normally effectively deployed against biotrophs. A key outcome of this is a loss of host cell membrane integrity which results in a flood of previously unobtainable nutrients into the apoplast where the fungus resides.12 Within this context, an incompatible interaction would require the as yet unidentified Avr-R protein interaction to directly or indirectly prevent these events occurring. The model suggests that M. graminicola produces plant PCD and MAPK activating toxins and/or effectors and that wheat leaf cells possess molecular targets for these factors. These “sensitivity factors” could even conceivably include HR triggering resistance protein homologues.2 Clearly this model requires further testing. However, one point is already becoming clear. It is perhaps time for a greater appreciation of the emerging mechanistic sophistication associated with successful plant infection by necrotrophic pathogens.
Figure 1.
Models illustrating the events that occur in wheat leaf cells during susceptible and resistant interactions with the necrotrophic fungal pathogen, Mycosphaerella graminicola. The models suggest that specific fungal toxin(s)/effector(s) targets are present in a healthy plant cell, along with relatively large amounts of the TaMPK6 protein when compared to TaMPK3 protein levels (indicated by different font sizes). During a susceptible disease interaction, approximately nine days post inoculation (dpi), inverse changes in the relative levels of the two MAPK proteins are observed. At this stage the fungus is stimulated to produce, as yet un-identified, toxin(s) and/or effector(s) that trigger the post-translational activation (**) of TaMPK3. These events occur in parallel with the activation of programmed cell death (PCD) signalling, which may also include the generation of reactive oxygen species (ROS). The net effect is a loss of host membrane integrity and the release of nutrients from dying plant cells which supports increased fungal growth and asexual reproduction. None of these responses occur during a resistant interaction. The possible sites for protective function of corresponding Avr-R protein combinations are indicated as parallel crossed lines. The net outcome of the lack of plant cell reactions during host resistance is a limitation on the nutrients available to the necrotrophic fungus which prevents its further proliferation.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/6292
References
- 1.De Wit PJG. Visions & reflections (minireview)—How plants recognize pathogens and defend themselves. Cellular and Molecular Life Sciences. 2007;64:2726–2732. doi: 10.1007/s00018-007-7284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorang JM, Sweat TA, Wolpert TJ. Plant disease susceptibility conferred by a “resistance” gene. Proc Natl Acad Sci USA. 2007;104:14861–14866. doi: 10.1073/pnas.0702572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg JT, Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 4.Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- 5.Jin HL, Liu YD, Yang KY, Kim CY, Baker B, Zhang SQ. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 2003;33:719–731. doi: 10.1046/j.1365-313x.2003.01664.x. [DOI] [PubMed] [Google Scholar]
- 6.Pedley KF, Martin GB. Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J Biol Chem. 2004;47:49229–49235. doi: 10.1074/jbc.M410323200. [DOI] [PubMed] [Google Scholar]
- 7.Romeis T, Piedras P, Zhang SQ, Klessig DF, Hirt H, Jones JDG. Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound and salicylate responses. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stulemeijer IJE, Stratmann JW, Joosten M. Tomato mitogen-activated protein kinases LeMPK1, LeMPK2 and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 2007;144:1481–1494. doi: 10.1104/pp.107.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SQ, Klessig DF. Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc Natl Acad of Sci USA. 1998;95:7433–7438. doi: 10.1073/pnas.95.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brading PA, Verstappen ECP, Kema GHJ, Brown JKM. A gene-for-gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology. 2002;92:439–445. doi: 10.1094/PHYTO.2002.92.4.439. [DOI] [PubMed] [Google Scholar]
- 11.Flor HH. Current Status of Gene-for-Gene Concept. Annu Rev Phytopathol. 1971;9:275–276. [Google Scholar]
- 12.Keon J, Antoniw J, Carzaniga R, Deller S, Ward JL, Baker JM, Beale MH, Hammond-Kosack KE, Rudd JJ. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death of its susceptible wheat host. Mol Plant-Microbe Interact. 2007;20:178–193. doi: 10.1094/MPMI-20-2-0178. [DOI] [PubMed] [Google Scholar]
- 13.Rudd JJ, Keon J, Hammond-Kosack KE. The wheat mitogen-activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 2008;147:802–815. doi: 10.104/pp.108.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Rudd JJ, Macioszek VK, Scheel D. Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem. 2004;279:22440–22448. doi: 10.1074/jbc.M401099200. [DOI] [PubMed] [Google Scholar]
- 16.Yang KY, Liu Y, Zhang S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA. 2001;98:741–746. doi: 10.1073/pnas.98.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang SQ, Seguin A, Ellis BE. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Shetty NP, Kristensen BK, Newman MA, Møller K, Gregersen PL, Jørgensen HJL. Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol Mol Plant Pathol. 2003;62:333–346. [Google Scholar]