Abstract
Tissue culture of immortal cell strains from diseased patients is an invaluable resource for medical research, but is largely limited to tumor cell lines or transformed derivatives of native tissues. Here we describe the generation of induced pluripotent stem (iPS) cells from patients with a variety of genetic diseases with either Mendelian or complex inheritance that include: adenosine deaminase deficiency-related severe combined immunodeficiency (ADA-SCID), Shwachman-Bodian-Diamond syndrome (SBDS), Gaucher disease (GD) type III, Duchenne (DMD) and Becker muscular dystrophy (BMD), Parkinson disease (PD), Huntington disease (HD), juvenile-onset, type 1 diabetes mellitus (JDM), Down syndrome (DS)/trisomy 21 and the carrier state of Lesch-Nyhan syndrome. Such patient-specific stem cells offer an unprecedented opportunity to recapitulate both normal and pathologic human tissue formation in vitro, thereby enabling disease investigation and drug development.
Introduction
Cell culture has been the back-bone of basic biomedical research for many decades, and countless insights into both normal and pathologic cellular processes have been gleaned by studying human cells explanted in vitro. Most of the human cell lines in wide use today carry genetic and epigenetic artifacts of accommodation to tissue culture, and are derived either from malignant tissues or are genetically modified to drive immortal growth (Grimm, 2004). Primary human cells have a limited lifespan in culture, a constraint that thwarts inquiry into the regulation of tissue formation, regeneration, and repair. Indeed, many human cell types have never faithfully been adapted for growth in vitro, and the lack of accessible models of normal and pathologic tissue formation has rendered many important questions in human development and disease pathogenesis inaccessible.
Human embryonic stem cells isolated from excess embryos from in vitro fertilization clinics represent an immortal propagation of pluripotent cells that theoretically can generate any cell type within the human body (Lerou et al., 2008; Murry and Keller, 2008). Human embryonic stem cells allow investigators to explore early human development through in vitro differentiation, which recapitulates aspects of normal gastrulation and tissue formation. Embryos shown to carry genetic diseases by virtue of preimplantation genetic diagnosis (PGD; genetic analysis of single blastomeres obtained by embryo biopsy) can yield stem cell lines that model single gene disorders (Verlinsky et al., 2005), but the vast majority of diseases that show more complex genetic patterns of inheritance are not represented in this pool.
A tractable method for establishing immortal cultures of pluripotent stem cells from diseased individuals would not only facilitate disease research, but also lay a foundation for producing autologous cell therapies that would avoid immune rejection and enable correction of gene defects prior to tissue reconstitution. One strategy for producing autologous, patient-derived pluripotent stem cells is somatic cell nuclear transfer (NT). In a proof of principle experiment, NT-ES cells generated from mice with genetic immunodeficiency were used to combine gene and cell therapy to repair the genetic defect (Rideout et al., 2002). To date, NT has not proven successful in the human, and given the paucity of human oocytes, is destined to have limited utility. In contrast, introducing a set of transcription factors linked to pluripotency can directly reprogram human somatic cells to produce induced pluripotent stem (iPS) cells, a method that has been achieved by several groups worldwide (Lowry et al., 2008; Park et al., 2008b; Takahashi et al., 2007; Yu et al., 2007). Given the robustness of the approach, direct reprogramming promises to be a facile source of patient-derived cell lines. Such lines would be immediately valuable for medical research, but current methods for reprogramming require infecting the somatic cells with multiple viral vectors, thereby precluding consideration of their use in transplantation medicine at this time.
Human cell culture is an essential complement to research with animal models of disease. Murine models of human congenital and acquired diseases are invaluable but provide a limited representation of human pathophysiology. Murine models do not always faithfully mimic human diseases, especially for human contiguous gene syndromes such as trisomy 21 (Down syndrome or DS). A mouse model for the DS critical region on distal human chromosome 21 fails to recapitulate the human cranial abnormalities commonly associated with trisomy 21 (Olson et al., 2004). Orthologous segments to human chromosome 21 are present on mouse chromosomes 10 and 17 and distal human chromosome 21 corresponds to mouse chromosome 16 where trisomy 16 in the mouse is lethal (Nelson and Gibbs, 2004). Thus, a true murine equivalent of human trisomy 21 does not exist. Murine strains carrying the same genetic deficiencies as the human bone marrow failure disease Fanconi anemia demonstrate DNA repair defects consistent with the human condition (e.g. (Chen et al., 1996), yet none develop the spontaneous bone marrow failure that is the hallmark of the human disease.
For cases where murine and human physiology differ, disease-specific pluripotent cells capable of differentiation into the various tissues affected in each condition could undoubtedly provide new insights into disease pathophysiology by permitting analysis in a human system, under controlled conditions in vitro, using a large number of genetically-modifiable cells, and in a manner specific to the genetic lesions in each - whether known or unknown. Here, we report the derivation of human iPS cell lines from patients with a range of human genetic diseases.
Results and Discussion
Dermal fibroblasts or bone marrow-derived mesenchymal cells were obtained from patients with a prior diagnosis of a specific disease, and used to establish disease-specific lines of human iPS cells (Table 1). This initial cohort of cell lines was derived from patients with Mendelian or complex genetic disorders, including: Down syndrome (DS; trisomy 21); adenosine deaminase deficiency-related severe combined immunodeficiency (ADA-SCID); Shwachman-Bodian-Diamond syndrome (SBDS); Gaucher disease (GD) type III; Duchenne type (DMD) and Becker type (BMD) muscular dystrophy; Huntington chorea (Huntington disease; HD); Parkinson disease (PD); juvenile-onset, type 1 diabetes mellitus (JDM); and Lesch-Nyhan syndrome (LNSc; carrier state).
Table 1.
iPS cells derived from somatic cells of patients with genetic disease
| Name | Disease | Molecular Defect | Donor cell | Age | Sex |
|---|---|---|---|---|---|
| ADA | ADA-SCID | GGG->AGG, exon 7 and Del(GAAGA) exon 10, ADA gene | fibroblast | 3 M | Male |
| GD | Gaucher disease type III | AAC->AGC, exon 9, and G-insertion, nucleotide 84 of cDNA, GBA gene | fibroblast | 20 Y | Male |
| DMD | Duchenne muscular dystrophy | Deletion of exon 45–52, dystrophin gene | fibroblast | 6 Y | Male |
| BMD | Becker muscular dystrophy | Unidentified mutation in dystrophin | fibroblast | 38 Y | Male |
| DS1, DS2 | Down Syndrome | Trisomy 21 | fibroblast | 1 Y, 1 M | Male |
| PD | Parkinson disease | Multifactorial | fibroblast | 57 Y | Male |
| JDM | Juvenile diabetes mellitus | Multifactorial | fibroblast | 42 Y | Female |
| SBDS | Shwachman-Bodian-Diamond syndrome | IV2+2T->C and IVS3-1G>A, SBDS gene | bone marrow mesenchymal cells | 4 M | Male |
| HD | Huntington disease | 72 CAG repeats, huntingtin gene | fibroblast | 20 Y | Female |
| LNSc | Lesch-Nyhan Syndrome (carrier) | Heterozygosity of HPRT1 | fibroblast | 34 Y | Female |
Patient-derived somatic cells were transduced with either four (OCT4, SOX2, KLF4, and c-MYC) or three reprogramming factors (lacking c-MYC). Following two to three weeks of culture in hES cell supporting conditions, compact refractile ES-like colonies emerged amongst a background of fibroblasts, as previously described (Park et al., 2008a; Park et al., 2008b). Although our previous report used additional factors (hTERT and SV40 LT) to achieve reprogramming of adult somatic cells, we have found the four-factor cocktail to be sufficient as long as we employ a higher multiplicity of retroviral infection. Additionally, we generated a single line from a carrier of Lesch-Nyhan Syndrome using five doxycycline-inducible lentiviral vectors (OCT4, SOX2, KLF4, c-MYC, and NANOG) a strategy that has been used to isolate murine iPS cells (Brambrink et al., 2008; Stadtfeld et al., 2008), but previously had not been attempted with human somatic cells. Characterization of the iPS lines is presented below.
Mutation analysis in iPS lines
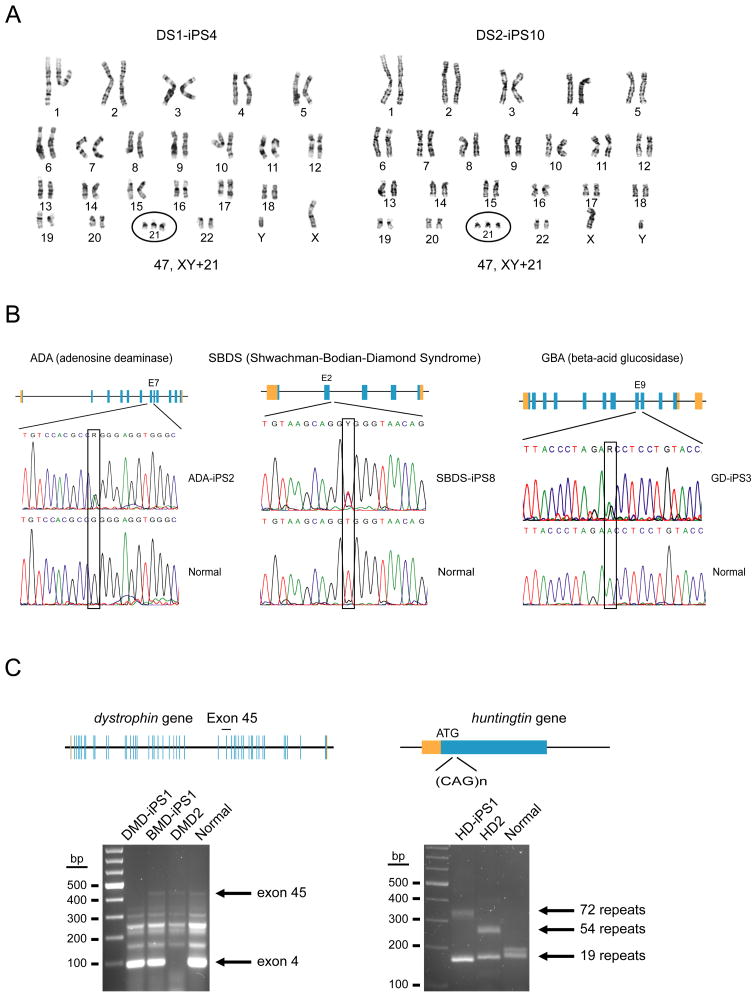
The iPS lines were evaluated to confirm, where possible, the disease-specific genotype of their parental somatic cells. Analysis of the karyotype of iPS lines derived from two individuals with Down syndrome showed the characteristic trisomy 21 anomaly (Figure 1A). Aneuploidies such as that occurring in DS are unambiguously associated with advanced maternal age (reviewed in Antonarakis et al., 2004) and as such, are occasionally detected in the preimplantation embryo when IVF is coupled with PGD. While it is possible that a discarded IVF embryo found to have trisomy 21 could be donated to attempt hES cell derivation, it is important to point out that many gestating DS embryos do not survive the prenatal period. Some studies place the frequency of spontaneous fetal demise (miscarriage) in DS to be above 40% (Bittles et al., 2007). Thus, the derivation of a human iPS line with trisomy 21 from an existing individual may be preferable, as such a line is most likely to harbor the complex genetic and epigenetic modifiers that favor full term gestation, and by virtue of the often lengthy medical history, will be a more informative resource for correlative clinical research.
Figure 1. Genotypic analysis of disease-specific iPS cell lines.
A) Two different, primary fibroblast specimens, DS1 and DS2 from male patients with Down syndrome (trisomy 21) were used to derive DS1-iPS4 and DS2-iPS10. Each has a 47, XY+21 karyotype over several passages (G-banding analysis). B) Fibroblast (ADA and GBA) or bone marrow mesenchymal cells (SBDS) were used to generate iPS lines. Mutated alleles identical to the original specimens were verified by DNA sequencing. Adenosine deaminase deficiency line ADA-iPS2, a compound heterozygote: GGG to GAA double transition in exon 7 of one allele (G216R substitution); the second allele is an exon 10 frame-shift deletion (-GAAGA) (Hirschhorn et al., 1993). Shwachman-Bodian-Diamond syndrome line SBDS-iPS8 is also a compound heterozygote: point mutations at the IV2+2T>C intron 2 splice donor site and an IVS3-1G>A mutation of the SBDS gene (Austin et al., 2005). GD-iPS3 (Gaucher disease type III); a 1226A>G point mutation (N370S substitution) and a guanine insertion at nucleotide 84 of the cDNA (84GG) (Beutler et al., 1991). C) Fibroblasts from patients diagnosed with either Duchenne (DMD) or Becker type muscular dystrophy (BMD): DMD-iPS1 has a deletion over exons 45–52 (multiplex PCR for the dystrophin gene). We could not determine a deletion in BMD-iPS1 using two different multiplex PCR sets though these assays do not cover the entire coding region. DMD2 is a patient control (exon 4 deletion). The control is genomic DNA from a healthy volunteer. Huntington disease (HD) is caused by a tri-nucleotide repeat expansion within the huntingtin locus. DNA sequencing shows that HD-iPS has one normal (<35 repeats) and one expanded allele (72 repeats). HD2 is a positive control from a second Huntington patient with one normal and one expanded allele (54 repeats). The control is genomic DNA from a healthy volunteer.
Creation of iPS lines from patients with single-gene disorders allows experiments on disease phenotypes in vitro, and an opportunity to repair gene defects ex vivo. The resulting cells, by virtue of their immortal growth in culture, can be extensively characterized to ensure that gene repair is precise and specific, thereby reducing the safety concerns of random, viral-mediated gene therapy. Repair of gene defects in pluripotent cells provides a common platform for combined gene repair and cell replacement therapy for a variety of genetic disorders, as long as the pluripotent cells can be differentiated into relevant somatic stem cell or tissue populations.
Three diseases in our cohort of iPS cells are inherited in a classical Mendelian manner as autosomal recessive congenital disorders, and are caused by point mutations in genes essential for normal immunologic and hematopoietic function: adenosine deaminase deficiency, which causes severe combined immune deficiency (ADA-SCID) due to the absence of T-cells, B-cells, and NK-cells; Shwachman-Bodian-Diamond syndrome, a congenital disorder characterized by exocrine pancreas insufficiency, skeletal abnormalities, and bone marrow failure; and Gaucher disease type III, an autosomal recessive lysosomal storage disease characterized by pancytopenia and progressive neurological deterioration due to mutations in the acid beta-glucosidase (GBA) gene. Sequence analysis of the ADA gene in the disease-associated ADA-iPS2 line revealed a compound heterozygote: a GGG to GAA transition mutation at exon 7, causing a G216R amino acid substitution (Figure 1B); the other allele is known to have a frame-shift deletion (-GAAGA) in exon 10 (Hirschhorn et al., 1993). The SBDS-iPS8 line harbors point mutations at the IV2+2T>C intron 2 splice donor site (Figure 1B) and IVS3-1G>A mutation (Austin et al., 2005). Molecular analysis of the GBA gene in the Gaucher disease line revealed a 1226A>G point mutation, causing a N370S amino acid substitution (Figure 1B); the second allele is known to have a frame-shifting insertion of a single guanine at cDNA nucleotide 84 (84GG) (Beutler et al., 1991). The Lesch-Nyhan syndrome carrier line harbors heterozygous deficiency of the HPRT gene (Nussbaum et al., 1983).
Two lines were derived from dermal fibroblasts cultured from patients with muscular dystrophy. Multiplex PCR analysis with primer sets amplifying several (but not all) intragenic intervals of the dystrophin gene (Beggs et al., 1990; Chamberlain et al., 1988) revealed the deletion of exons 45–52 in the iPS cells derived from a patient with Duchenne muscular dystrophy (DMD; Figure 1C). Despite analysis for gross genomic defects by multiplex PCR, a deletion was not detected in iPS cells derived from a patient with Becker type muscular dystrophy (BMD; Figure 1C). As BMD is a milder form of disease, and the dystrophin gene one of the largest in the human genome, definition of the genetic lesion responsible for this condition is sometimes elusive (Prior and Bridgeman, 2005).
Given that numerous groups have pioneered the directed differentiation of neuronal subtypes, and that genetically defined ES cells from animal models of amyotrophic lateral sclerosis have revealed important insights into the pathophysiology of motor neuron deterioration (Di Giorgio et al., 2007), there is considerable interest in generating iPS lines from patients afflicted with neurodegenerative disease. We generated iPS lines from a patient with Huntington chorea (Huntington disease; HD), and verified the presence of expanded (CAG)n polyglutamine triplet repeat sequences (72) in the proximal portion of the huntingtin gene (Figure 1C; (Riess et al., 1993) in one allele and 19 repeats in the other (where the normal range is 35 or less (Chong et al., 1997).
Pluripotent cell lines will likewise be valuable for studying neurodegenerative conditions with more complex genetic predisposition, as well as metabolic diseases known to have familial predispositions but for which the genetic contribution remains unexplained. We have generated lines from a patient diagnosed with Parkinson disease and another from a patient with juvenile onset (Type I) diabetes mellitus (Table 1). Given that these conditions lack a defined genetic basis, genotypic verification is impossible at this time.
The Lesch-Nyhan syndrome is caused by mutations in Hypoxanthine-guanine phosphoribosyltransferase (HPRT), an X-linked enzyme in purine metabolism that when deficient leads to abnormal accumulation of uric acid and a neurologic disorder characterized by cognitive deficits and self-mutilating behavior. Cells carrying either intact or deficient HPRT enzyme function can be selectively cultured in media containing Hypoxanthine-Aminopterin-Thymidine (HAT) or 6-thioguanine (6-TG), respectively. Strategies for inducing specific mutation or gene repair by homologous recombination were first established for the HPRT locus (Doetschman et al., 1987; Doetschman et al., 1988; Thomas and Capecchi, 1987). We have generated an iPS line from a female carrier (LNSc-iPS2) that will be a valuable resource for studies of homologous recombination in iPS cells, and for analysis of X chromosome reactivation during reprogramming and random inactivation with differentiation.
Characterization of disease-related iPS lines
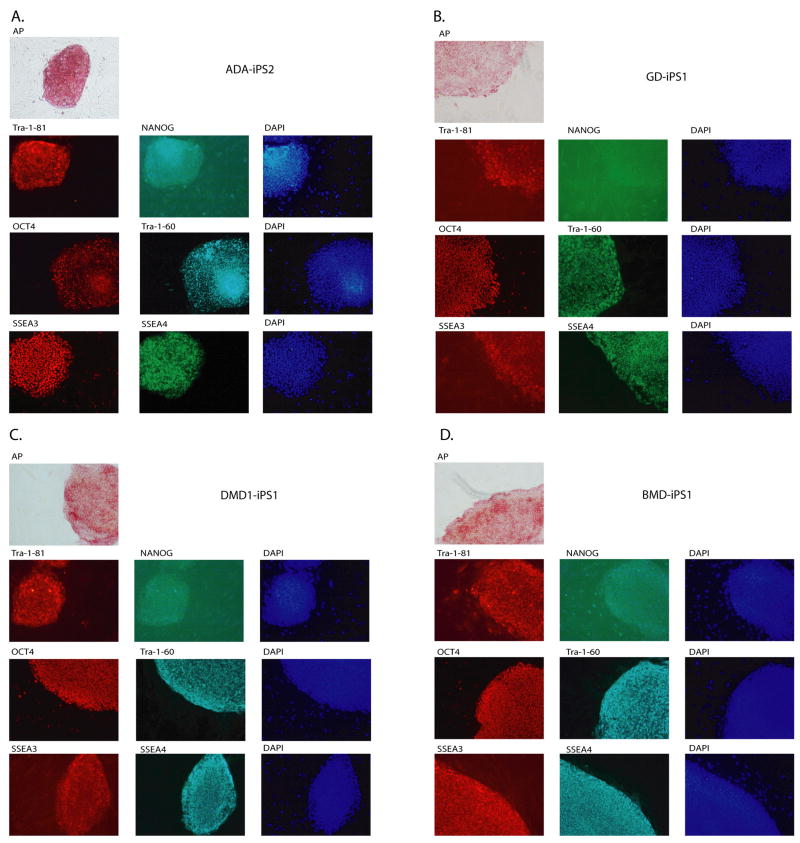
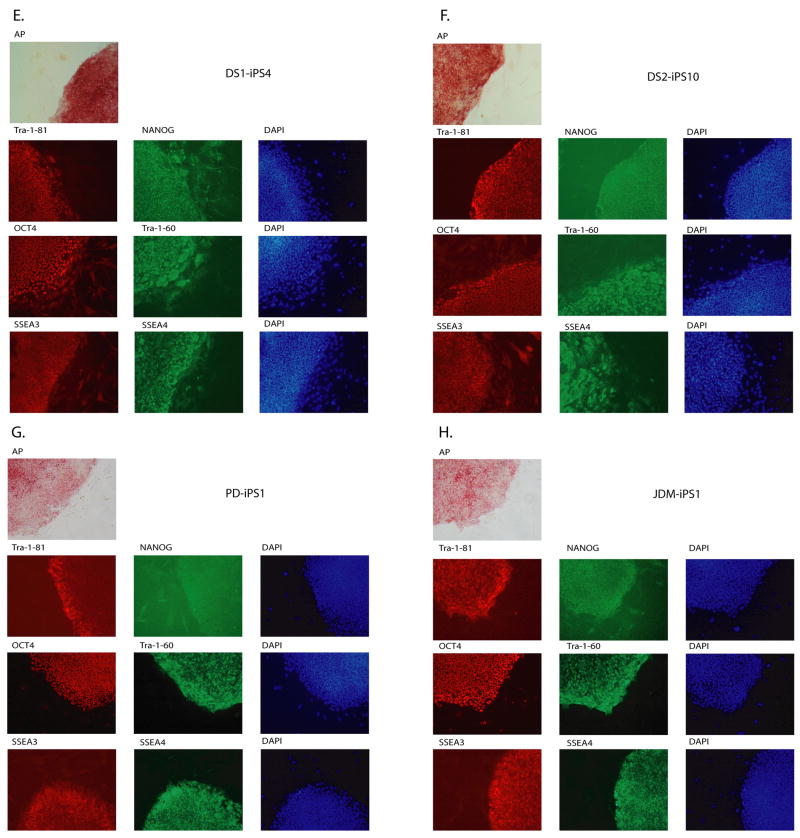
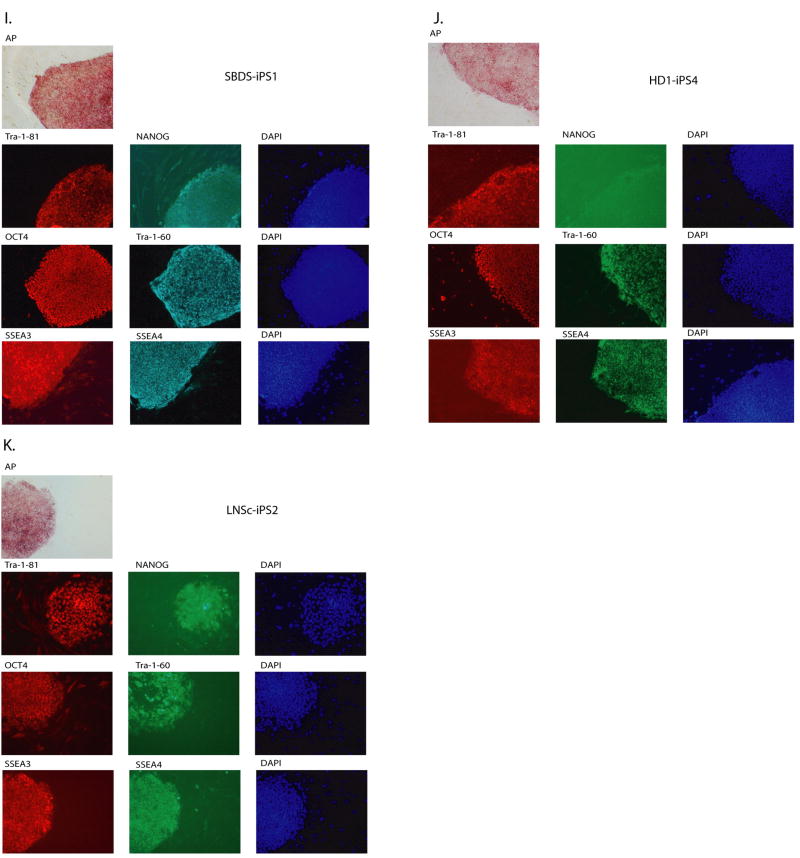
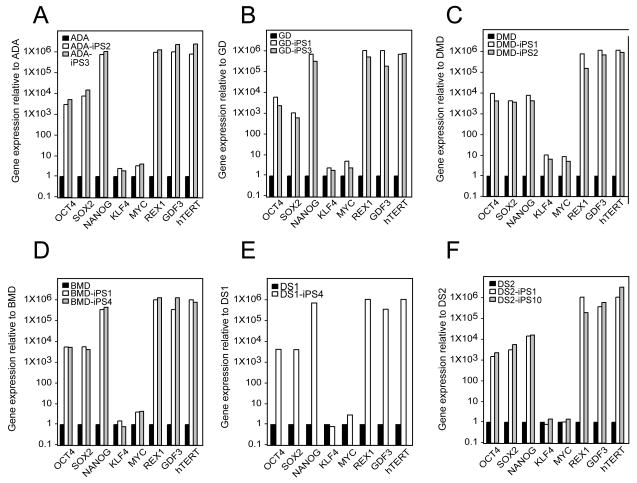
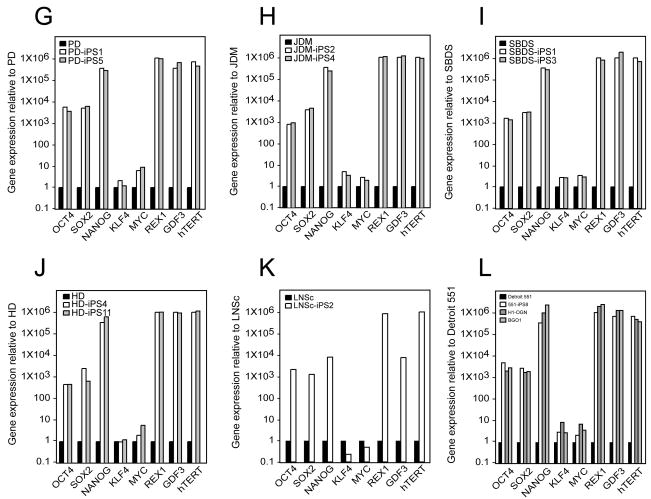
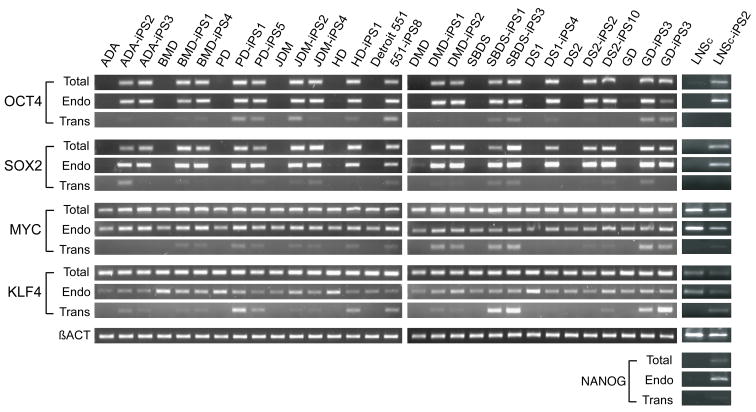
All iPS colonies, which were selected based on their morphologic resemblance to colonies of ES cells, demonstrated compact colony morphology and markers of pluripotent cells, including alkaline phosphatase (AP), Tra-1-81, Tra-1-60, OCT4, NANOG, SSEA3 and SSEA4 (Figure 2). Quantitative RT-PCR indicated the expression of pluripotency-related genes including OCT4, SOX2, NANOG, REX1, GDF3, and hTERT regardless of the genetic condition represented within the parental somatic cells (Figure 3; control lines are shown in panel l). Retroviral transgenes were largely silenced in the iPS lines, with expression of the relevant reprogramming factors assumed by endogenous loci (Figure 4), as described (Park et al., 2008b). PCR-based DNA fingerprint analysis using highly-variable number of tandem repeats (VNTR) confirmed that the iPS lines were genetically matched to their parental somatic lines, ruling out the possibility of cross-contamination from existing cultures of human pluripotent cells (Supplemental Figure 1). Also, iPS cells showed normal 46 XX, or 46 XY karyotypes (Supplementary Figure 2).
Figure 2. Patient-derived iPS lines exhibit markers of pluripotency.
ADA-iPS2, GD-iPS1, DMD-iPS1, BMD-iPS1, DS1-iPS4, DS2-iPS10, PD-iPS1, JDM-iPS1, SBDS-iPS1, HD-iPS4, LNSc-iPS2, JDM-iPS2 were established from a fibroblast or mesenchymal cells (Table 1). Disease specific iPS cell lines maintain a morphology similar to hES cells when grown in co-culture with mouse embryonic feeder fibroblasts (MEFs). Patient-specific iPS cells express alkaline phosphatase (AP). Also, as shown here via immunohistochemistry, patient-specific cells express pluripotency markers including Tra-1-81, NANOG, OCT4, Tra-1-60, SSEA3 and SSEA4. 4,6-Diamidino-2-phenylindole (DAPI) staining is shown at right and indicates the total cell content per image.
Figure 3. Expression of pluripotency-associate genes is elevated in patient-specific iPS lines relative to their somatic cell controls.
In each panel, quantitative real-time PCR (QRT-PCR) assays for OCT4, SOX2, NANOG, REX1, GDF3, and hTERT indicates increased expression in patient-specific iPS cells relative to parent cell lines while expression of KLF4 and cMYC remains largely unchanged. PCR reactions were normalized against internal controls (β-actin) and plotted relative to expression levels in their individual parent fibroblast cell lines. (A) the human iPS lines ADA-iPS2 and –iPS3 are derived from the adenosine deaminase deficiency-severe combined immunodeficiency fibroblast line ADA. (B) GD-iPS1 and –iPS3 are derived from the Gaucher disease type III fibroblast line GD. (C) DMD-iPS1 and –iPS2 are derived from the Duchenne muscular dystrophy fibroblast line DMD. (D) BMD-iPS1 and –iPS4 are derived from the Becker muscular dystrophy line BMD. (E) DS1-iPS4 is derived from the Down syndrome fibroblast line DS1. (F) DS2-iPS1 and –iPS10 are derived from the Down syndrome fibroblast line DS2. (G) PD-iPS1 and –iPS5 are derived from the Parkinson disease fibroblast line PD. (H) JDM-iPS2 and –iPS4 are derived from the juvenile-onset, type 1 diabetes mellitus line JDM. (I) SBDS-iPS1 and –iPS3 are derived from the Shwachman-Bodian-Diamond syndrome bone marrow mesenchymal fibroblast line SBDS. (J) HD-iPS4 and –iPS11 are derived from the Huntington disease fibroblast line HD. (K) LNSc-iPS1 and –iPS2 are derived from the Lesch-Nyhan syndrome carrier fibroblast line LNSc. (L) Detroit 551 human fibroblasts are used as the standard here in order to demonstrate the previously described expression pattern in Detroit 551 derived iPS cells (551-iPS8) relative to two bona fide hES cell lines: H1-OGN and BG01.
Figure 4. Pluripotency-promoting genes are chiefly expressed from the endogenous loci in patient-specific iPS lines, while the virally-delivered transgene is predominantly silenced.
The patient-specific iPS cell lines shown here are preceded by their parental fibroblast controls (from left to right at top): adenosine deaminase deficiency-associate severe combined immunodeficiency (ADA), Becker muscular dystrophy (BMD), Parkinson disease (PD), juvenile type one diabetes mellitus (JDM), Huntington disease (HD), Detroit 551 control cells, Duchenne muscular dystrophy (DMD), Shwachman-Bodian-Diamond syndrome (SBDS), Down syndrome (DS), Gaucher disease type III (GD), and Lesch-Nyhan syndrome carrier (LNSc). The semi-quantitative expression (RT-PCR) of the four pluripotency-promoting genes used in the reprogramming process, OCT4, SOX2, cMYC, KLF4 and NANOG is shown for each line using amplification conditions specific to the endogenous (Endo) or virally-delivered transgene (Trans) as well as the total expression for each (Total). Beta-actin is shown at the bottom as a loading control for each lane.
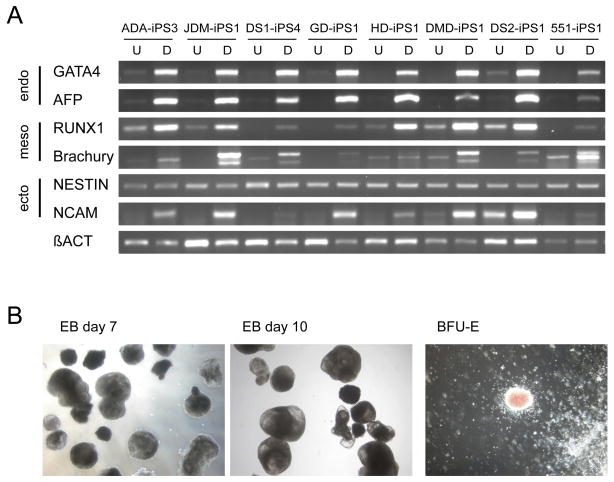
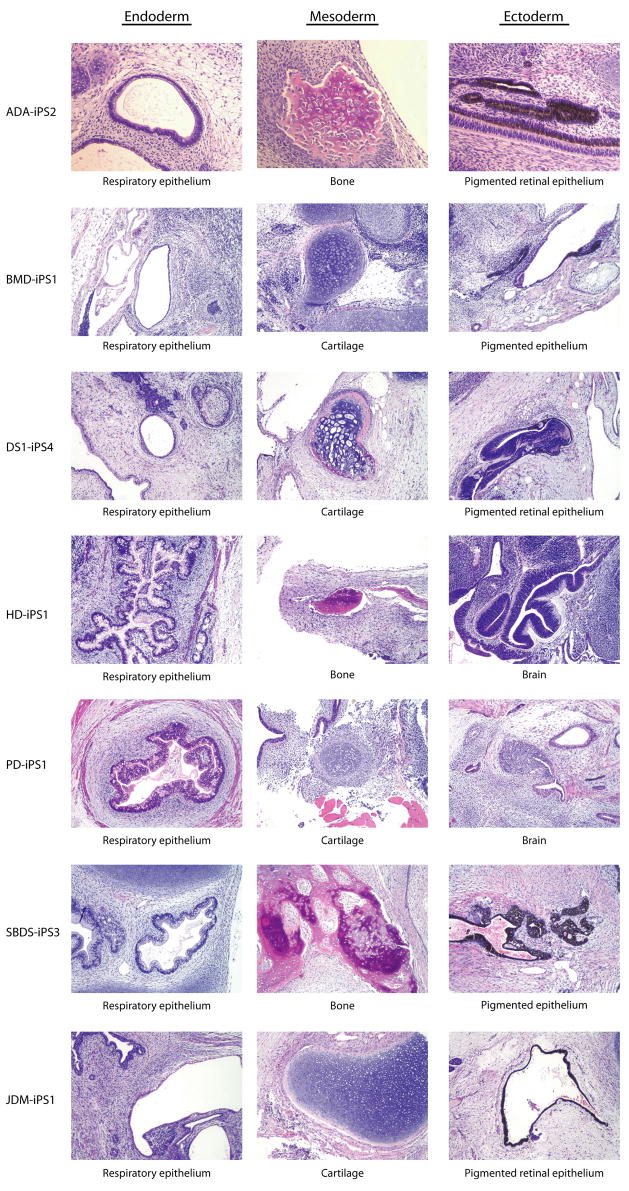
Human disease-associated iPS lines were characterized by a standard set of assays to confirm pluripotency and multi-lineage differentiation. iPS lines (n=7) were allowed to differentiate in vitro into embryoid bodies as described (Park et al., 2008b), and their potential to develop along specific lineages was confirmed by PCR for markers of all three embryonic germ layers (ectoderm, mesoderm, and endoderm; Supplemental Figure 6a). Hematopoietic differentiation of disease-specific iPS lines (n=2) produced myeloid and erythroid colony types (Figure 5). The ultimate standard of pluripotency for human cells is teratoma formation in immunodeficient murine hosts (Lensch et al., 2007). When injected sub-cutaneously into immunodeficient Rag2−/−γC−/− mice, disease-specific iPS lines (n=7) produced mature, cystic masses representing all three embryonic germ layers (Figure 6).
Figure 5. Differentiation of patient-specific iPS lines reveals lineage-specific gene expression and mature cell formation.
A) At top (from left to right) are nine iPS cell lines in their undifferentiated (U) or differentiated (D) state. The lines are: adenosine deaminase deficiency-associated severe combined immunodeficiency (ADA), juvenile-onset type one diabetes mellitus (JDM), Down syndrome 1 (DS1), Gaucher disease type III (GD), Huntington disease (HD), Duchenne muscular dystrophy (DMD), Down syndrome 2 (DS2), and normal control Detroit 551 (551) cells. Differentiation (D) of these patient-specific iPS cells as embryoid bodies (EB) followed by RT-PCR analysis shows upregulated expression of lineage markers from the three embryonic germ layers relative to their undifferentiated controls (U) including: GATA4 and AFP (endoderm), RUNX1 and Brachyury (mesoderm), and Nestin and NCAM (ectoderm). Beta-actin serves as a positive amplification control for each. B) Differentiation of ADA-iPS2, a representative patient-specific iPS cell line, as embryoid bodies (EB) is highly reminiscent of that using hES cells where tight clusters of differentiating cells are well-formed by day 7 which will cavitate, becoming cystic, by day 10. Hematopoietic differentiation of patient-specific iPS cells yields various blood cell types in semi-solid methylcellulose colony-forming assays including burst-forming unit-erythroid (BFU-E) which are derivative of red blood cell progenitor cells.
Figure 6. Patient-specific iPS lines form teratomas in immunodeficient mice.
Shown here are the representative series of hematoxylin-eosin (H/E) stained sections from a formalin fixed teratoma produced from ADA-iPS2, BMD-iPS1, DS1-iPS4, HD-iPS1, PD-iPS1, SBDS-iPS3, and JDM-iPS1 cell lines. They formed mature, cystic teratomas with tissues representing all three embryonic germ layers including: respiratory epithelium (endoderm), bone and cartilage (mesoderm), and pigmented retinal epithelium and immature neural tissue (ectoderm).
The technique of factor-based reprogramming of somatic cells generates pluripotent stem cell lines that are effectively immortal in culture and can be differentiated into any of a multitude of human tissues. By comparison of normal and pathologic tissue formation, and by assessment of the reparative effects of drug treatment in vitro, cell lines generated from patients offer an unprecedented opportunity to recapitulate pathologic human tissue formation in vitro, and a new technology platform for drug screening. The Harvard Stem Cell Institute has committed resources to establish a Core Facility for the production of disease-specific iPS lines, with the goal of making each of these lines available to the biomedical research community.
Experimental Procedures
Somatic cell culture, isolation and culture of iPS cells
Fibroblasts from patients with ADA-SCID (ADA, GM01390), Gaucher disease (GD, GM00852), Duchenne type muscular dystrophy (DMD, GM04981; DMD2, GM05089), Becker type muscular dystrophy (BMD, GM04569), Down syndrome (DS1, AG0539A), Parkinson disease (PD, AG20446), juvenile (Type I) diabetes mellitus (JDM, GM02416), Huntington disease (HD, GM04281; HD2, GM01187), and Lesch-Nyhan syndrome carrier (LNSc, GM00013) were obtained from Coriell. Fibroblasts from patients with Down syndrome (DS2, DLL54) and normal fetal skin fibroblasts (Detroit 551) were purchased from ATCC. Bone marrow mesenchymal cells from SBDS patient (SBDS, DF250) has been described (Austin et al., 2005). Cells were grown in alpha-MEM containing 10 % inactivated fetal serum (IFS), 50 U/ml penicillin, 50 mg/ml streptomycin, and 1 mM L-glutamine. Retroviruses expressing OCT4, SOX2, KLF4, and MYC were pseudotyped in VSVg and used to infect 1×105 cells in one well of a six-well dish. iPS cells were isolated as described previously (Park et al., 2008b). iPS cells from LNSc fibroblasts were isolated using an inducible lentiviral system as previously described (Stadtfeld et al, 2008). cDNAs encoding human OCT4, SOX2, cMYC, KLF4, and NANOG were cloned into doxycycline inducible vectors and were co-infected with a lentivirus harboring a constitutively expressed reverse tetracycline transactivator (rtTA). Infected fibroblasts were split to feeders under hES culture conditions. Doxycycline was added to the culture for 30 days, and then withdrawn. Colonies that appeared were picked and expanded into lines in the absence of doxycycline. iPS colonies were maintained in hES medium (80% DMEM/F12, 20% KO Serum Replacement,10 ng/ml bFGF, 1 mM L-glutamine, 100 μM nonessential amino acids, 100 μM 2-mercaptoethanol, 50 U/ml penicillin, and 50 mg/ml streptomycin).
Characterization of genetic defects in iPS cells
Genomic DNA was isolated from cells using DNeasy kit (Qiagen). PCR reactions were performed using 50 ng of genomic DNA with primers corresponding to the mutated regions of genes responsible for each condition (ADA-SCID, Gaucher disease, SBDS (Calado et al., 2007), and Huntington disease). Primers sequences are provided in Supplementary Table 1. PCR products were resolved via agarose gels, purified and sequenced, or cloned into the TOPO vector (Invitrogen) for sequencing. The number of CAG repeats in the HD gene was determined by amplifying the 5− end of the huntingtin gene by PCR and sequencing. The deletion of exons within the dystrophin gene in DMD-iPS cells and BMD-iPS cells was determined by PCR using Chamberlain or Beggs’ multiplex primer sets (Beggs et al., 1990; Chamberlain et al., 1988).
Karyotype analysis
Chromosomal studies including karyotype of trisomy 21 in DS1-iPS and DS2-iPS10 cells were performed at the Cytogenetics Core of the Dana-Farber/Harvard Cancer Center or Cell Line Genetics using standard protocols for high-resolution G-banding.
Fingerprinting analysis
50 ng of genomic DNA was used to amplify across discrete genomic intervals containing highly variable numbers of tandem repeats (VNTR). PCR products were resolved in 3 % agarose gels to examine the differential amplicon mobility for each primer set: D10S1214, repeat (GGAA)n, average heterozygosity 0.97; D17S1290, repeat (GATA)n, average heterozygosity 0.84; D7S796, repeat (GATA)n, average heterozygosity 0.95; and D21S2055, repeat (GATA)n, average heterozygosity 0.88 (Invitrogen).
Immunohistochemistry and AP staining of iPS cells
iPS cells grown on feeder cells were fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 for 30 minutes, and blocked in 3% BSA in PBS for 2 hours. Cells were incubated with primary antibody overnight at 4 °C, washed, and incubated with Alexa Fluor (Invitrogen) secondary antibody for 3 hours. SSEA-3, SSEA-4, TRA 1–60, TRA 1–81 antibodies were obtained from Millipore. OCT3/4 and NANOG antibodies were obtained from Abcam. Alkaline phosphatase staining was done per the manufacturer’s recommendations (Millipore).
Analysis of gene expression
Total RNA was isolated from iPS cells using an RNeasy kit (Qiagen) according to the manufacturer’s protocol. 0.5 μg of RNA was subjected to the RT reaction using Superscript II (Invitrogen). Quantitative PCR was performed with Brilliant SYBR Green Master MiX in Stratagene MX3000P machine using previously described primers (Park et al., 2008b). Semi-quantitative PCR was performed to look at the expression of total, endogenous and recombinant pluripotency genes, and genes representing the three embryonic germ layers using primers described previously and in Supplementary Table 1.
Differentiation of iPS cells
iPS cells were washed with DMEM/F12, treated with collagenase for 10 min, and collected by scraping. Colonies were washed once with DMEM/F12, and gently resuspended in EB differentiation medium. EBs were differentiated with low-speed shaking and the medium was changed every three days. After two weeks of differentiation, EBs were dissociated and plated in MethoCult (Stem Cell Technologies).
Teratoma formation from iPS cells
iPS cells were washed with DMEM/F12, treated with collagenase for 10 min at room temperature, scraped using glass pipette, and collected by centrifugation. Cells were washed once with DMEM/F12, and mixed with Matrigel (BD Biosciences) and collagen (Sigma). 2×106 cells were intramuscularly injected into immune deficient Rag2−/−γC−/− mice. After 6 weeks of injection, teratomas were dissected, rinsed once with PBS, and fixed in 10% formalin. Embedding in paraffin, sectioning of tissue, and Hematoxylin/Eosin staining were performed by the Rodent Histopathology service of the Dana Farber Cancer Institute.
Supplementary Material
Supplemental Figure 1. Qualitative DNA fingerprint analysis indicates that each line is derivative of its indicated parental fibroblast source.
PCR-based DNA fingerprint analysis using primer sets spanning highly variable tetra-nucleotide repeats are shown for four different loci: D7S796, repeat (GATA)n, average heterozygosity 0.95; D21S2055, repeat (GATA)n, average heterozygosity 0.88; D17S1290, repeat (GATA)n, average heterozygosity 0.84; and D10S1214, repeat (GGAA)n, average heterozygosity 0.97. Of note, the Down syndrome derived iPS lines (DS1-iPS4 and DS2-iPS3) as well as their respective parent fibroblasts (DS1 and DS2) each show three alleles at D21S2055 in keeping with the observation that most cases of DS derive from errors occurring within meiosis I of female germ cell development, where the two maternal amplicons represent alleles from each maternal grandparent with the third allele originating from within the paternal genome. From left to right at top are six lines of previously described (Park et al., 2008) human iPS cells: MRC5-iPS2 is a normal iPS line from fetal lung fibroblasts, BJ1-iPS4 is a normal iPS line from neonatal foreskin fibroblasts, MSC-iPS2 is a normal iPS line from mesenchymal fibroblasts, hFib2-iPS2 is a normal iPS line from adult fibroblasts, and 551-iPS8 is a normal fibroblast iPS line. These are followed (from left to right) by patient-specific iPS lines as well as their parental fibroblast controls: DMD = Duchenne muscular dystrophy, SBDS = Shwachman-Bodian-Diamond syndrome, DS = Down syndrome, GD = Gaucher disease type III, ADA = adenosine deaminase deficiency-associated severe combined immunodeficiency, BMD = Becker type muscular dystrophy, PD = Parkinson disease, JDM = juvenile-onset type one diabetes mellitus, HD = Huntington disease, LNSc = Lesch-Nyhan syndrome carrier, H1-OGN and BG01 are human embryo-derived hES cells, and 293T is an immortalized human embryonic kidney-derived cell line used in the creation of the viral supernatants for reprogramming.
Supplemental Figure 2. Patient-specific iPS lines maintain normal karyotypes.
When chromosomal contents were analyzed with high resolution G-banding karyotypes, ADA-iPS3, GD-iPS1, DMD-iPS1, BMD-iPS4, PD-iPS5, JDM-iPS1, SBDS-iPS3, and HD-iPS1 indicated normal, diploid chromosomal contents.
Acknowledgments
GQD was supported by grants from the National Institutes of Health, the NIH Director’s Pioneer Award of the NIH Roadmap for Medical Research, and private funds contributed to the Harvard Stem Cell Institute and the Children’s Hospital Stem Cell Program. GQD is a recipient of Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukemia and Lymphoma Society, and is an Investigator of the Howard Hughes Medical Institute. KH was supported by the NIH Director’s Innovator Award and the Harvard Stem Cell Institute. NM was supported by the Natural Sciences and Engineering Council of Canada and a Sir James Lougheed Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Austin KM, Leary RJ, Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs AH, Koenig M, Boyce FM, Kunkel LM. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86:45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Kuhl W, Sorge J, West C. Identification of the second common Jewish Gaucher disease mutation makes possible population-based screening for the heterozygous state. Proc Natl Acad Sci U S A. 1991;88:10544–10547. doi: 10.1073/pnas.88.23.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Bower C, Hussain R, Glasson EJ. The four ages of Down syndrome. Eur J Public Health. 2007;17:221–225. doi: 10.1093/eurpub/ckl103. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado RT, Graf SA, Wilkerson KL, Kajigaya S, Ancliff PJ, Dror Y, Chanock SJ, Lansdorp PM, Young NS. Mutations in the SBDS gene in acquired aplastic anemia. Blood. 2007;110:1141–1146. doi: 10.1182/blood-2007-03-080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- Chong SS, Almqvist E, Telenius H, LaTray L, Nichol K, Bourdelat-Parks B, Goldberg YP, Haddad BR, Richards F, Sillence D, et al. Contribution of DNA sequence and CAG size to mutation frequencies of intermediate alleles for Huntington disease: evidence from single sperm analyses. Hum Mol Genet. 1997;6:301–309. doi: 10.1093/hmg/6.2.301. [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- Doetschman T, Maeda N, Smithies O. Targeted mutation of the Hprt gene in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. The art and design of genetic screens: mammalian culture cells. Nat Rev Genet. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R, Chen AS, Israni A, Yang DR, Huie ML. Two new mutations at the adenosine deaminase (ADA) locus (Q254X and del nt1050–54) unusual for not being missense mutations. Hum Mutat. 1993;2:320–323. doi: 10.1002/humu.1380020415. [DOI] [PubMed] [Google Scholar]
- Lensch MW, Schlaeger TM, Zon LI, Daley GQ. Teratoma formation assays with human embryonic stem cells: a rationale for one type of human-animal chimera. Cell Stem Cell. 2007;1:253–258. doi: 10.1016/j.stem.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Lerou PH, Yabuuchi A, Huo H, Takeuchi A, Shea J, Cimini T, Ince TA, Ginsburg E, Racowsky C, Daley GQ. Human embryonic stem cell derivation from poor-quality embryos. Nat Biotechnol. 2008;26:212–214. doi: 10.1038/nbt1378. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Gibbs RA. Genetics. The critical region in trisomy 21. Science. 2004;306:619–621. doi: 10.1126/science.1105226. [DOI] [PubMed] [Google Scholar]
- Nussbaum RL, Crowder WE, Nyhan WL, Caskey CT. A three-allele restriction-fragment-length polymorphism at the hypoxanthine phosphoribosyltransferase locus in man. Proc Natl Acad Sci U S A. 1983;80:4035–4039. doi: 10.1073/pnas.80.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008a;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Prior TW, Bridgeman SJ. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J Mol Diagn. 2005;7:317–326. doi: 10.1016/S1525-1578(10)60560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Hochedlinger K, Kyba M, Daley GQ, Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- Riess O, Noerremoelle A, Soerensen SA, Epplen JT. Improved PCR conditions for the stretch of (CAG)n repeats causing Huntington’s disease. Hum Mol Genet. 1993;2:637. doi: 10.1093/hmg/2.6.637. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, Kuliev A. Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Qualitative DNA fingerprint analysis indicates that each line is derivative of its indicated parental fibroblast source.
PCR-based DNA fingerprint analysis using primer sets spanning highly variable tetra-nucleotide repeats are shown for four different loci: D7S796, repeat (GATA)n, average heterozygosity 0.95; D21S2055, repeat (GATA)n, average heterozygosity 0.88; D17S1290, repeat (GATA)n, average heterozygosity 0.84; and D10S1214, repeat (GGAA)n, average heterozygosity 0.97. Of note, the Down syndrome derived iPS lines (DS1-iPS4 and DS2-iPS3) as well as their respective parent fibroblasts (DS1 and DS2) each show three alleles at D21S2055 in keeping with the observation that most cases of DS derive from errors occurring within meiosis I of female germ cell development, where the two maternal amplicons represent alleles from each maternal grandparent with the third allele originating from within the paternal genome. From left to right at top are six lines of previously described (Park et al., 2008) human iPS cells: MRC5-iPS2 is a normal iPS line from fetal lung fibroblasts, BJ1-iPS4 is a normal iPS line from neonatal foreskin fibroblasts, MSC-iPS2 is a normal iPS line from mesenchymal fibroblasts, hFib2-iPS2 is a normal iPS line from adult fibroblasts, and 551-iPS8 is a normal fibroblast iPS line. These are followed (from left to right) by patient-specific iPS lines as well as their parental fibroblast controls: DMD = Duchenne muscular dystrophy, SBDS = Shwachman-Bodian-Diamond syndrome, DS = Down syndrome, GD = Gaucher disease type III, ADA = adenosine deaminase deficiency-associated severe combined immunodeficiency, BMD = Becker type muscular dystrophy, PD = Parkinson disease, JDM = juvenile-onset type one diabetes mellitus, HD = Huntington disease, LNSc = Lesch-Nyhan syndrome carrier, H1-OGN and BG01 are human embryo-derived hES cells, and 293T is an immortalized human embryonic kidney-derived cell line used in the creation of the viral supernatants for reprogramming.
Supplemental Figure 2. Patient-specific iPS lines maintain normal karyotypes.
When chromosomal contents were analyzed with high resolution G-banding karyotypes, ADA-iPS3, GD-iPS1, DMD-iPS1, BMD-iPS4, PD-iPS5, JDM-iPS1, SBDS-iPS3, and HD-iPS1 indicated normal, diploid chromosomal contents.