Abstract
The Arabidopsis (Arabidopsis thaliana) Heavy Metal Associated3 (AtHMA3) protein belongs to the P1B-2 subgroup of the P-type ATPase family, which is involved in heavy metal transport. In a previous study, we have shown, using heterologous expression in the yeast Saccharomyces cerevisiae, that in the presence of toxic metals, AtHMA3 was able to phenotypically complement the cadmium/lead (Cd/Pb)-hypersensitive strain ycf1 but not the zinc (Zn)-hypersensitive strain zrc1. In this study, we demonstrate that AtHMA3 in planta is located in the vacuolar membrane, with a high expression level in guard cells, hydathodes, vascular tissues, and the root apex. Confocal imaging in the presence of the Zn/Cd fluorescent probe BTC-5N revealed that AtHMA3 participates in the vacuolar storage of Cd. A T-DNA insertional mutant was found more sensitive to Zn and Cd. Conversely, ectopic overexpression of AtHMA3 improved plant tolerance to Cd, cobalt, Pb, and Zn; Cd accumulation increased by about 2- to 3-fold in plants overexpressing AtHMA3 compared with wild-type plants. Thus, AtHMA3 likely plays a role in the detoxification of biological (Zn) and nonbiological (Cd, cobalt, and Pb) heavy metals by participating in their vacuolar sequestration, an original function for a P1B-2 ATPase in a multicellular eukaryote.
Plant metal homeostasis must be tightly regulated to ensure sufficient micronutrient (Zn, Cu, Fe, etc.) supply to the different organs and to prevent toxic concentrations of these and nonessential metals (Cd, Pb, Co, etc.) from inducing deleterious effects (Clemens, 2001; Fraustro da Silva and Williams, 2001; Hall, 2002). Transporters belonging to various additional families, such as Cation Diffusion Facilitator (CDF), Natural resistance-associated macrophage protein (Nramp), ATP Binding Cassette (ABC), Zinc Iron-like Protein (ZIP), and others, have already been shown to be involved in these processes (for review, see Colangelo and Guerinot, 2006; Clemens, 2006; Krämer et al., 2007), and this list is quickly extending. One of the most recent additions has been a member of the major facilitator superfamily, ZIF1 (for Zinc Induced Facilitator), for its participation in Zn/Fe homeostasis (Haydon and Cobbett, 2007).
The P1B-ATPase subfamily HMA (for Heavy Metal Associated) plays an important role in the process of metal allocation or detoxification (Williams and Mills, 2005). CadA from Staphylococcus aureus or Listeria monocytogenes (Lebrun et al., 1994) and ZntA from Escherichia coli (Rensing et al., 1997; Sharma et al., 2000) belong to the Zn/Cd/Pb subgroup and are among the first P1B-ATPases that have been functionally characterized; they are implicated in the detoxification of the bacteria by an ATP-dependent efflux of metals. Among the 46 genes identified in the genome of Arabidopsis (Arabidopsis thaliana) to encode P-type ATPases, eight belong to the group of HMAs, HMA1 to HMA8, based on previously proposed nomenclature (Baxter et al., 2003). This subfamily clusters in two subgroups depending on heavy metal specificity: Cu+/(Ag+) or Zn2+/Cd2+/Pb2+/Co2+; AtHMA5 to -8 and AtHMA1 to -4 belong to those subgroups, respectively. The copper transporter family has been studied earlier in planta (Hirayama et al., 1999; Woeste and Kieber, 2000; Shikanai et al., 2003; Abdel-Ghany et al., 2005; Andrés-Colas et al., 2005; Puig et al., 2007). AtHMA1 was listed in the P1B-4 subgroup (Argüello, 2003), a cluster including CoaT, a cobalt efflux pump from Synechocystis (Rutherford et al., 1999). In planta, AtHMA1 is expressed at the inner envelope membrane of the chloroplast and participates in the loading of copper into the stroma (Seigneurin-Berny et al., 2006).
Thus, based on amino acid alignments (Argüello, 2003), the Zn2+/Cd2+/Pb2+/Co2+ subgroup in Arabidopsis potentially includes three members, AtHMA2, AtHMA3, and AtHMA4. These proteins present a high degree of amino acid sequence similarities in their transmembrane domains, but they display divergent soluble C-terminal segments. AtHMA2 and AtHMA4 present unusually long C-terminal sequences (around 250 and 460 residues, respectively), with many Cys doublets and ending with a His stretch. In contrast, the AtHMA3 C terminus is shorter (around 60 residues) and includes only two Cys doublets. In planta, AtHMA4 is expressed at the plasma membrane, and studies of T-DNA insertional mutants have demonstrated that this protein is involved in Zn and Cd xylem loading and in the translocation of these metals from the roots to the shoot (Hussain et al., 2004; Verret et al., 2004). Interestingly, the plant tolerance to Zn/Cd was enhanced in AtHMA4-overexpressing plants (Verret et al., 2004). The functional characterization of AtHMA2 in planta has demonstrated a large redundancy with AtHMA4 in Zn transport (Eren and Argüello, 2004; Hussain et al., 2004). A hma2 hma4 double mutant had a strong Zn nutritional deficiency and was sterile due to an absence of pollen. These results demonstrate that AtHMA4 and AtHMA2 proteins are the main transporters that ensure Zn translocation from roots to the shoot (Hussain et al., 2004). Heterologous expression of the AtHMA3 cDNA from the Wassilewskija (Ws) ecotype in yeast led us to propose a role of this protein in Cd/Pb transport (Gravot et al., 2004). AtHMA3 expression was able to rescue the Cd/Pb-hypersensitive phenotype of the mutant strain ycf1 but unable to phenotypically complement the Zn-sensitive mutant zrc1. Various transcriptomic studies have shown that AtHMA3 mRNA is present at a very weak level in all plant parts and that its expression is only slightly modulated by Zn or Cd treatments, if affected at all (Becher et al., 2004; Gravot et al., 2004; Talke et al., 2006). In contrast, the ortholog of AtHMA3 in the tolerant hyperaccumulator species Arabidopsis halleri is strongly expressed and even up-regulated upon Zn exposure (Becher et al., 2004; Gravot et al., 2004; Talke et al., 2006). These observations suggest that HMA3 could be one of the transporters implicated in metal hyperaccumulation and/or metal tolerance processes. AtHMA3 is a pseudogene in the wild-type Columbia (Col-0) ecotype (Hussain et al., 2004). A single base pair deletion at position 1,626 of the Col-0 cDNA (position in relation to the ATG start codon) results in a frameshift, leading to a premature stop codon and a subsequent truncated protein (Supplemental Fig. S1). Additionally, a T-DNA insertional mutant in the Ws background, Athma3-1, was found to be indistinguishable from the wild type under normal culture conditions (Hussain et al., 2004).
Our study deals with the functional characterization of AtHMA3 in planta in the Ws background. AtHMA3 is found to be a vacuolar transporter whose overexpression results in Cd, Pb, Cd, and Zn tolerance.
RESULTS
HMA3 Localizes at the Vacuolar Membrane
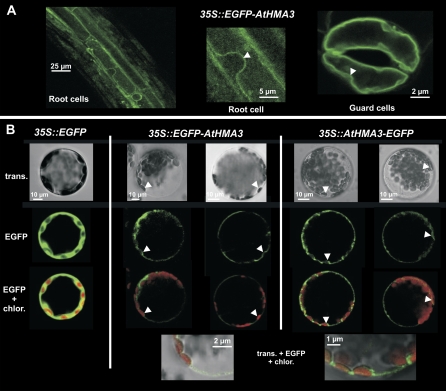
To determine the subcellular location of AtHMA3, an N-terminal EGFP fusion was obtained by cloning the EGFP cDNA in frame with the AtHMA3 cDNA under the control of the cauliflower mosaic virus 35S promoter (CaMV35S). Ws plants were transformed with this clone, and four homozygous, independent strains were obtained by hygromycin B selection (13C, 18J, 19K, and 27G). The subcellular localization of the EGFP∷AtHMA3 fusion protein was determined by confocal microscopy. In addition to the cell wall autofluorescence, the green fluorescence of the EGFP fusion protein was observed at the level of the tonoplast, delineating the nucleus, in root cells, and guard cells (Fig. 1A). Since the fusion of the EGFP could lead to a mistargeting of the protein, two EGFP fusion proteins (EGFP∷AtHMA3 and AtHMA3∷EGFP) were also transiently expressed in mesophyll protoplasts from Arabidopsis. When the plasma membrane and the tonoplast were separated by the nucleus or chloroplasts, the fluorescent pattern of the two fusion proteins was clearly consistent with a localization of AtHMA3 at the vacuolar membrane (Fig. 1B).
Figure 1.
AtHMA3 localizes at the tonoplast. Confocal imaging of the expression patterns of the EGFP∷AtHMA3 and AtHMA3∷EGFP chimeric protein fusions. A, Root cells and guard cells from plants expressing the EGFP∷AtHMA3 fusion protein. B, Transient expression in mesophyll protoplasts of EGFP (left), EGFP∷AtHMA3 (middle), and AtHMA3∷EGFP (right). Arrows indicate regions where the tonoplast is separated from the plasma membrane.
AtHMA3 Expression Pattern as Revealed by GUS Activity
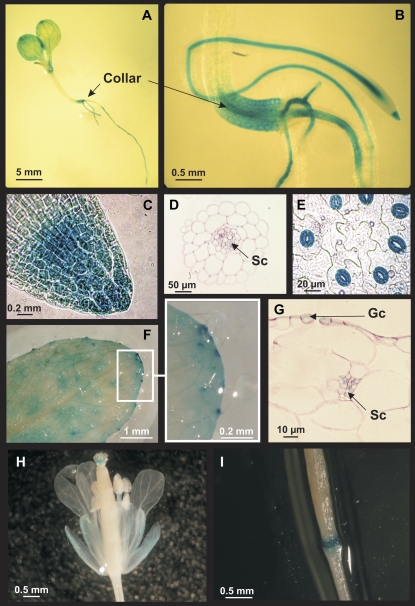
GUS activity was used to determine AtHMA3 expression pattern in planta. The promoter region of AtHMA3, 2,747 bp upstream of the ATG start codon, was fused with the uidA gene, which encodes GUS; the fusion gene was expressed in transgenic Ws plants. Three homozygous independent lines were obtained by selection on hygromycin B. GUS expression was mainly observed in root, collar, and leaf cells of 4- to 6-d-old plantlets (Fig. 2, A and B). In roots, GUS activity was seen in vascular bundles and at the apex (Fig. 2, B and C). In a root section, staining was restricted to stellar cells around the vascular vessels (Fig. 2D). In leaves, GUS was strongly expressed in guard cells (Fig. 2, E and G) and hydathodes (Fig. 2F). In a leaf cross-section, GUS staining was also present in vascular bundles (Fig. 2G). In inflorescence, the expression was observed in stigma, stamen filaments, and vascular tissues of sepals (Fig. 2H) as well as at the abscission zone of siliques (Fig. 2I). This expression profile is consistent with data compiled from various microarray experiments (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). The GUS staining pattern and intensity were not modified when plantlets were incubated in the presence of 200 μm Zn or 30 μm Cd (data not shown).
Figure 2.
AtHMA3 is expressed in several plant tissues. Expression profile of AtHMA3 in planta revealed by GUS activity. A, Whole plantlet. B, Detail from A. C, Root apex. D, Root section. E, Leaf epidermis. F, Leaf. G, Leaf section. H, Inflorescence. I, Silique. Gc, Guard cell; Sc, stellar cells.
An Athma3 T-DNA Insertional Mutant Is Zn and Cd Sensitive
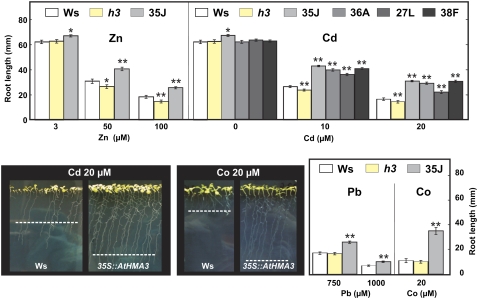
A T-DNA insertional mutant of AtHMA3, Athma3-1, has already been described in the Ws background (Hussain et al., 2004). The T-DNA is inserted in the first exon, and no AtHMA3 transcript was found in this mutant, which did not exhibit any particular phenotype under control conditions (Hussain et al., 2004). Zn, Cd, Pb, and Co metal tolerance of this line was estimated by measuring the root length of plantlets, which were grown vertically on solid bactoagar medium. In control conditions, at 3 μm Zn in the nutrient solution, Athma3-1 showed as much development as the wild-type seedlings (Fig. 3). While there was no difference when this line was challenged with Pb or Co, the metal tolerance to Zn and Cd was slightly impaired in the mutant line compared with the wild type (Fig. 3).
Figure 3.
AtHMA3 enhances heavy metal tolerance of Arabidopsis plantlets. Root length measurements of wild-type (Ws), Athma3-1 (h3), and 35S∷AtHMA3- or 35S∷EGFP∷AtHMA3-overexpressing seedlings. Plantlets were vertically grown in the presence of the indicated metal concentrations. Photographs of plates and root length measurements were done 14 d after germination. The dashed lines on the photographs represent the mean values of root lengths. Values are means of 100 to 150 root length measurements ± se, representative of three independent experiments. Significant differences from the wild type as determined by Student's t test are indicated by one (P < 0.05) or two (P < 0.001) asterisks.
AtHMA3 Overexpression Enhances Plant Metal Tolerance
Plants ectopically overexpressing AtHMA3 were generated by agrotransformation of Arabidopsis strain Ws with the AtHMA3 cDNA cloned under the control of the constitutive promoter CaMV35S. Four independent 35S∷EGFP∷AtHMA3 lines (13C, 18J, 19K, and 27G) and four 35S∷AtHMA3 lines (35J, 36A, 27L, and 38F), which represented the higher amounts of AtHMA3 transcripts, were selected (see semiquantitative reverse transcription [RT]-PCR in Supplemental Fig. S2). All experiments presented below were performed on wild-type Ws and the overexpressing lines. Zn, Cd, Pb, and Co metal tolerance of these lines was estimated as described above (Fig. 3; Supplemental Fig. S3). In the presence of toxic concentrations of Zn, 50 and 100 mm, the mean root length of the wild-type plantlets was reduced by 50% and 72%, respectively (Fig. 3). Under the same conditions, the overexpressing plantlets exhibited 34% and 39% longer roots than the wild-type plants. The tolerance to Cd, Pb, and Co in the 35S∷AtHMA3 lines was also tested. In all cases, root lengths of the plants overexpressing AtHMA3 were less affected by the heavy metals than those of the wild-type plants (Fig. 3).
AtHMA3 Overexpression Induces an Increased Accumulation of Cd in Plants
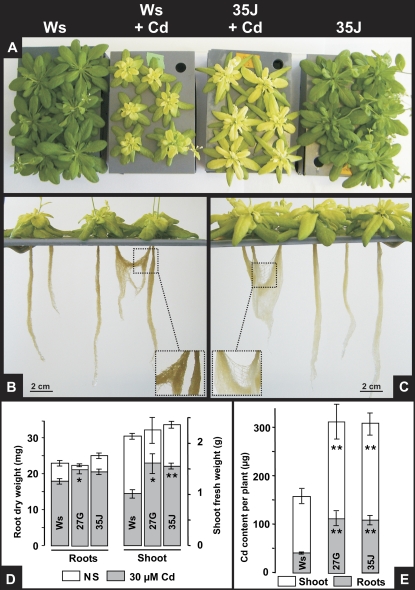
In order to test the effect that ectopic expression of AtHMA3 has on plant tolerance to Cd and metal accumulation, hydroponic cultures of wild-type and overexpressing plants were performed for 2 weeks in nutrient solution, followed by 11 d in nutrient solution, supplemented or not with a 30 μm Cd solution. The Cd treatment led to typical chlorotic symptoms in all lines, but the overexpressors exhibited a better tolerance to the toxic metal, which was characterized by a higher growth rate. The roots of wild-type plants became brown in the presence of Cd (Fig. 4, B and C), as reported previously (Howden et al., 1995a, 1995b). Such intense brownish coloration was seen in roots of the glutathione-deficient mutants cad1 and cad2, which are unable to accumulate and sequester Cd. This brown coloration was faint in the overexpressors, suggesting that these plants were less stressed than the wild-type plants (Fig. 4, C compared with B). In accordance, overexpressing lines exhibited better shoot development than the wild-type plants by about 50% (Fig. 4D). At the end of the Cd treatment, metal contents in the roots and the shoot of the different lines were determined by inductively coupled plasma-atomic emission spectrometry (ICP-AES) analyses. Compared with wild-type plants, both overexpressing lines studied, 27G and 35J, were found to have accumulated about 2.5-fold more Cd in the roots and about 2-fold in the shoots (Fig. 4E).
Figure 4.
AtHMA3 overexpression in planta enhances Cd tolerance and accumulation. A, Plants were grown hydroponically in the aerated nutrient solution, in the presence or absence of 30 μm Cd, for 11 d. The nutrient solution and, when present, the Cd solution were renewed every 2 d. B and C, Roots of wild-type Ws plants (B) exhibited a brownish coloration less perceptible in the roots of the overexpressor lines 27G and 35J (C). D and E, Leaves and roots were harvested, weighed (D), dried, and mineralized, and the metal content was determined by ICP-AES measurements (E). Values are means of 12 measurements for each line ± se. Significant differences from the wild-type as determined by Student's t test are indicated by one (P < 0.05) or two (P < 0.001) asterisks.
AtHMA3 Mediates Cd Sequestration into the Vacuole
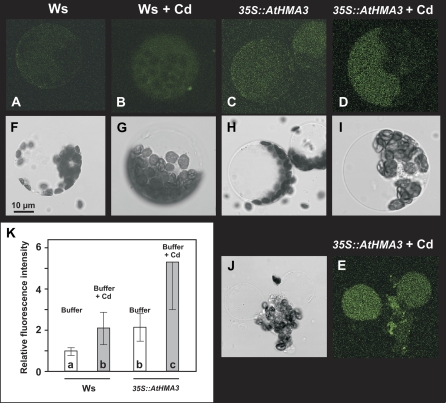
Due to AtHMA3 localization at the vacuolar membrane, the role of this transporter in heavy metal sequestration into the vacuole was investigated. Cd uptake in the vacuole of leaf protoplasts was followed using a Cd/Zn fluorescent probe, the acetoxymethyl ester form of the fluoroprobe BTC-5N/AM (acetoxymethyl ester of 5-nitrobenzothiazole coumarin). Protoplasts, loaded with the fluorophore, were incubated in the presence of 0.35 mm Cd, a concentration found optimal for fluorescence recovery in wheat (Triticum aestivum) protoplasts (Lindberg et al., 2004). The fluorescence exhibited by protoplasts, from the wild-type Ws and the overexpressing 35S∷AtHMA3 lines, was analyzed in the presence and absence of Cd (Fig. 5). A low signal was detected in wild-type protoplasts; this could be attributed to the intracellular Zn content (Fig. 5A; Supplemental Movie S1). The addition of 0.35 mm Cd increased the level of fluorescence in wild-type protoplasts (Fig. 5B; Supplemental Movie S2). The signal was observed throughout the cell, with the exception of the chloroplasts, suggesting that Cd entered the cytoplasm and reached the vacuole. In the absence of Cd, protoplasts from the 35S∷AtHMA3 lines exhibited an enhanced fluorescence compared with wild-type protoplasts (Fig. 5C; Supplemental Movie S3). After incubation in the presence of 0.35 μm Cd, a strong fluorescence was observed inside the vacuole of overexpressing 35S∷AtHMA3 protoplasts (Fig. 5D; Supplemental Movie S4). Treatment with a hypotonic medium led to a burst of the protoplasts, leading to highly fluorescent isolated vacuoles (Fig. 5E; Supplemental Movie S4).
Figure 5.
AtHMA3 participates in the metal sequestration into the vacuole. Protoplasts from wild-type and 35S∷AtHMA3-overexpressing plants were loaded with BTC-5N/AM. A to E, Green fluorescence of Zn- or Cd-conjugated BTC-5N. F to J, Corresponding transmission images. A and F, The control untreated protoplasts of Ws plants. B and G, The Ws protoplasts incubated with 0.35 μm Cd. C and H, The control untreated protoplasts of 35S∷AtHMA3 plants. D and I, The 35S∷AtHMA3 protoplasts incubated with 0.35 μm Cd. E and J, The same as D and I, after plasmolysis of the protoplasts. K, Fluorescence quantification, performed on eight to 12 protoplasts for each genotype and condition, using ImageJ 1.36 (http://rsb.info.nih.gov/ij/). Statistical treatment of the results was done using Kruskal-Wallis ANOVA on ranks (Sigmastat version 3.5); populations a, b, and c were significantly different (P < 0.05).
AtHMA3 Is a Pseudogene in Some Ecotypes
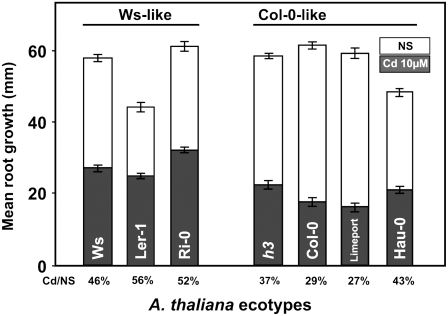
AtHMA3 is a pseudogene in the Col-0 strain (Hussain et al., 2004). The presence of a single base pair deletion at position 1,626 of the cDNA results in a frameshift, inducing a premature stop codon immediately downstream. The corresponding AtHMA3 mRNA putatively encodes a 542-amino acid polypeptide lacking the two last transmembrane helices and a large part of the cytosolic loop between the sixth and seventh helices, which present specific motifs implicated in the catalytic cycle. A 675-bp DNA fragment from 15 Arabidopsis ecotypes was amplified on the genomic DNA (Table I). After sequencing, it appeared that two of them (Limeport and Hau-0) exhibit the same base pair deletion as the Col-0 strain. Root growth experiments were carried out for Ws, Col-0, and two Ws-like and two Col-0-like ecotypes (Fig. 6). Under control conditions, the root length of the various lines was similar for Ws-like HMA3 ecotypes and Col-0-like ecotypes (P < 0.946). When the plants were grown under 10 μm Cd, the root lengths of all lines were shorter, but the effect was more obvious for Col-0 and Col-0-like ecotypes. The root development under Cd treatment was significantly higher in Ws-like ecotypes compared with Col-0-like ecotypes exhibiting the point mutation (P < 0.03). Similar results were obtained in a second independent experiment (data not shown).
Table I.
AtHMA3 status in various Arabidopsis ecotypes
The presence of a full-length AtHMA3 coding gene was studied for 15 Arabidopsis ecotypes. + indicates the presence of a full AtHMA3 coding sequence; − indicates the occurrence of a base pair deletion at position 1,626, relative to the ATG start codon.
| Accession Name | Original Stock Center No. | Geographic Origin | HMA3 Status |
|---|---|---|---|
| Be-0 | N964 | Bensheim, Germany | + |
| Cha-1 | N1070 | Champex, Switzerland | + |
| Col-0 | N22625 | Columbia, New York | − |
| Cvi-0 | N1096 | Green Cape Islands | + |
| Gr-2 | N1200 | Graz, Austria | + |
| Gre-0 | N1210 | Greenville, Mississippi | + |
| Hau-0 | N1221 | Hauniensis, Denmark | − |
| Hi-0 | N1227 | Hilversum, The Netherlands | + |
| Ler-1 | N22618 | Landsberg, Germany | + |
| Limeport | N8070 | Friedensville, Pennsylvania | − |
| Ost-0 | N1430 | Osthammer, Sweden | + |
| Pla-1 | N1460 | Playa de Oro, Spain | + |
| Ri-0 | N1492 | Richmond, British Columbia, Canada | + |
| Tol-0 | N8020 | Toledo, Ohio | + |
| Ws | N915 | Wassilewskija, Belarus | + |
Figure 6.
AtHMA3 participates in Cd tolerance in Arabidopsis. Root length in the presence of 10 μm Cd compared with that on nutrient solution for various Arabidopsis ecotype plantlets. The experiments were performed as described for Figure 3. Ler and Ri-0 ecotypes present a functional AtHMA3 like Ws, while Limeport and Hau-0 ecotypes are Col-0 like, which share a frameshift in the gene encoding AtHMA3. Values are means of 100 to 150 root length measurements ± se. All Col-0-like ecotypes were more sensitive to Cd than Ws-like ecotypes (P < 0.05). Statistical treatment of the results was done using Kruskal-Wallis ANOVA on ranks (Sigmastat version 3.5). NS, Nutrient solution (Zn concentration = 3 μm).
DISCUSSION
The P1B-ATPases are involved in heavy metal transport through biological membranes via an ATP-dependent process. While those participating in copper transport (P1B-1 subgroup) are present in all kingdoms (Axelsen and Palmgren, 1998), P1B-ATPases responsible for Zn, Cd, Pb, and Co transport (P1B-2 subgroup) are lacking in animals. In prokaryotes, this last subgroup plays an important physiological role in heavy metal detoxification. A prototypical example is CadA, conferring Cd tolerance to S. aureus through metal efflux (Lebrun et al., 1994). Eight P1B-ATPases (HMAs) are encoded in the Arabidopsis genome; three of them belong to the P1B-2-ATPase subgroup (AtHMA2 to -4) and are predicted as putative Zn/Cd/Pb/Co transporters. AtHMA2 and AtHMA4 have been extensively studied by means of heterologous expression, Arabidopsis knockout, and overexpressing mutant analyses (Eren and Argüello, 2004; Hussain et al., 2004; Verret et al., 2004, 2005). They both transport Zn, and in planta they are the main components for the long-distance transport of this micronutrient from the roots to the shoots (Hussain et al., 2004; Verret et al., 2004). In contrast, the physiological function of AtHMA3, the shortest P1B-2-ATPase, which is lacking the characteristic long C-terminal extension exhibited by AtHMA2 (Supplemental Fig. S1), and AtHMA4 was still unknown. This lack of knowledge may be due to the fact that AtHMA3 is a pseudogene in the main studied Arabidopsis ecotype, Col-0 (see below). In a previous study, it has been shown by means of RT-PCR that AtHMA3 was expressed elsewhere in the plant, but at a low level compared with AtHMA4 (Gravot et al., 2004). When expressed in yeast mutant strains, AtHMA3 was able to rescue the Cd/Pb-sensitive phenotype of the ycf1 strain; however, AtHMA3 was unable to complement the Zn-sensitive phenotype of the zrc1 strain (Gravot et al., 2004). These results pointed to a possible role of AtHMA3 in the compartmentalization of toxic heavy metals. In accordance, in this study we found that a T-DNA insertional mutant, Athma3-1, was slightly more sensitive to Cd and Zn at the level of root growth. The rather weak phenotype could result from the low expression level of the AtHMA3 gene in Arabidopsis.
AtHMA3 Subcellular Localization and Expression Pattern
To obtain further insight into AtHMA3 function, the subcellular localization of the protein was investigated in Ws plants expressing the GFP fused to AtHMA3. Confocal imaging showed that AtHMA3 is targeted to the vacuolar membrane (Fig. 1, A and B). This tonoplastic localization of AtHMA3 suggests a role of the protein in the transfer of heavy metals from the cytoplasm into the vacuole. This subcellular location contrasts with those previously determined for the other HMA proteins. Three proteins participate in chloroplastic copper homeostasis, AtHMA1 and AtHMA6, located at the chloroplast inner membrane (Shikanai et al., 2003; Abdel-Ghany et al., 2005; Seigneurin-Berny et al., 2006), and AtHMA8, at the thylakoid membrane (Abdel-Ghany et al., 2005). AtHMA7, a copper transporter, is located in the Golgi membrane (Hirayama et al., 1999; Woeste and Kieber, 2000), while AtHMA2 and AtHMA4, two Zn transporters, are embedded in the plasma membrane (Hussain et al., 2004; Verret et al., 2004). AtHMA5, which participates in copper detoxification, is putatively present at the plasma membrane (Andrés-Colas et al., 2005).
The expression pattern of the AtHMA3 gene has shown that it is expressed in the roots, in the stellar cells that surround the vascular vessels (Fig. 2, A, B, D, and G), and at the root apex (Fig. 2C). Previous RT-PCR analysis of the AtHMA3 level of expression (Gravot et al., 2004) and microarray transcript profiling on HMA3 from A. halleri (Becher et al., 2004; Talke et al., 2006) agree with the expression of AtHMA3 in the roots. At the shoot level, AtHMA3 is essentially expressed in the vasculature (Fig. 2A), in guard cells, and in hydathodes (Fig. 2, E and G). Altogether, the expression profile of AtHMA3 is reminiscent of that of AtHMA2 or AtHMA4 (Hussain et al., 2004; Verret et al., 2004), with the exception of guard cells, hydathodes, and the root apex. This suggests that these proteins may act in a cooperative way to maintain the cytoplasmic metal homeostasis in specific cells. AtHMA2 and AtHMA4, located at the plasma membrane, are involved in Zn translocation from roots to the shoot. AtHMA4, and likely AtHMA2, is able to load the xylem with Cd, Co, and Pb, which are toxic to the plant. We hypothesize that AtHMA3, expressed in the same cells, could act as a “filter” by decreasing the cytoplasmic concentration of toxic metals (Cd, Pb, and Co); this may be accomplished by sequestration into the vacuole and would limit the export of Cd, Pb, and Co or excess Zn to other tissues by AtHMA2/4. Additionally, AtHMA3 is highly expressed in the root tip, which may face the presence of heavy metals in the soil, in guard cells, and in hydathodes, which are located at the end of the transpiration stream, where heavy metals coming from the soil or from aerosols can accumulate after evaporation of the apoplastic solute. Many metal transporters that can promote a cytoplasmic Cd influx are expressed in guard cells (Leonhardt et al., 2004). The ZIP4 homolog in the hyperaccumulator species Thlapsi caerulescens is specifically expressed in guard cells (Küpper et al., 2007) and transports Cd and Zn (Pence et al., 2000). Additionally, it has been shown that Cd can penetrate the guard cells through hyperpolarized Ca2+ channels (Perfus-Barbeoch et al., 2002). In this context, a large activity of AtHMA3 could participate in cytoplasmic detoxification of Cd in guard cells, these cells being critical to maintain the plant water status. AtHMA3 expression is also observed in the inflorescence at the level of the filament of the stamen, the junction between the style and the stigma, and the abscission zone of the silique (Fig. 2, H and I).
AtHMA3 Overexpression Confers Heavy Metal Tolerance
The targeting of AtHMA3 to the vacuolar membrane suggests that it participates in heavy metal loading into vacuoles. A fluorescent Zn/Cd probe, BTC-5N, was used to visualize in vivo the intracellular Cd patterning, as already reported for wheat protoplasts (Lindberg et al., 2004). This probe has a roughly equal affinity for Cd and Zn, thus preventing discrimination between these two cations (Haugland, 1994). Nevertheless, in isolated protoplasts, a Cd treatment clearly enhanced the fluorescence intensity in comparison with untreated protoplasts. This increase was rather faint when wild-type Ws protoplasts were examined (Fig. 5, B compared with A), whereas the signal was largely enhanced when protoplasts overexpressing AtHMA3 were used (Fig. 5, D and E compared with C; Supplemental Movies S1–S4). After plasmolysis, the fluorescence was mostly found in the vacuole (Fig. 5E; Supplemental Movie S4); this coincides with the vacuolar localization of AtHMA3. These observations support that AtHMA3 plays a role in the cytoplasmic detoxification of the cells by pumping heavy metals into the vacuole and thus increasing plant tolerance. Constitutive ectopic expression of AtHMA3 in planta led to an increased tolerance to Co, Cd, Zn, and Pb. AtHMA3 could play a role in Zn compartmentalization and/or detoxification, but it is noteworthy that the improved metal tolerance was particularly obvious for the highly toxic nonessential metals, essentially Co and Cd (Figs. 3 and 4). Similar roles for P1B-2 ATPases in metal detoxification are well documented in prokaryotes (Lebrun et al., 1994; Rensing et al., 1997; Sharma et al., 2000), but, to our knowledge, this is an original observation of such a role in a multicellular eukaryote. The function of AtHMA3, in planta, would be somewhat homologous to that of YCF1 in Saccharomyces cerevisiae, participating in the vacuolar sequestration of toxic metals such as Cd and Pb (Li et al., 1996; Song et al., 2003).
Phytoremediation is an emerging biotechnology that uses plants to detoxify contaminated soils. This technique has the drawback of being time consuming, but there is a strong interest in increasing plant tolerance and metal translocation to improve the yield of metal recovery. Heavy metal tolerance and a high potential of metal transfer from roots to the shoot are common properties of metallophytes, metal-tolerant species that accumulate abnormally high contents of heavy metal in their leaves. A. halleri is a metallophyte species, close to Arabidopsis, and is able to accumulate high amounts of Zn and Cd. Comparison studies have shown that in this species, AhHMA3, likely orthologous to AtHMA3, is highly expressed, more than 100-fold the level found in Arabidopsis (Becher et al., 2004; Talke et al., 2006). Accordingly, overexpression of AtHMA3 in Arabidopsis led to more tolerant plants and to a higher metal content in the leaves (Fig. 4).
AtHMA3 Ecotypic Variability
AtHMA3 is nonfunctional in the Arabidopsis ecotype Col-0 (Hussain et al., 2004). In this accession, the cDNA presents a base pair deletion inducing a stop codon, producing a truncated protein that is lacking the essential large cytosolic loop and the seventh and eighth transmembrane helices. This may explain why various proteomic analyses performed on the tonoplast in the Col-0 background failed to detect AtHMA3 (Carter et al., 2004; Shimaoka et al., 2004; Jaquinod et al., 2007). Among 15 accessions, spanning the world, only three present this mutation. The ecotypes sharing the mutation were all found to be less tolerant of Cd, with some variability, than those exhibiting the full-length AtHMA3 cDNA (Fig. 6). This observation suggests a correlation between the presence of a functional AtHMA3 and heavy metal tolerance. A segregation analysis will be necessary to precisely determine the weight of AtHMA3 among other factors in plant tolerance to heavy metals.
In conclusion, we propose that AtHMA3 is involved in heavy metal tolerance in plants, along with other transporters such as CAX and metal chelators like phytochelatins, metallothioneins, and organic acids (Korenkov et al., 2007a, 2007b; for review, see Clemens et al., 2002). The role of the protein is to detoxify toxic metals by storing them in root cell vacuoles, which prevents this essential organ from poisoning. Moreover, it is interesting that ectopic overexpression of the protein favors the plant metal tolerance and accumulation, through metal sequestration in the vacuoles of aerial parts, a property that should be useful in phytoremediation.
MATERIALS AND METHODS
Plants
Arabidopsis (Arabidopsis thaliana ecotype Ws) plants were grown in a controlled environment (8-h photoperiod at 300 μmol m−2 s−1, 21°C, and 70% relative humidity) in a nutrient solution [800 μm Ca(NO3)2, 4H2O; 2 mm KNO3; 1.1 mm MgSO4, 7 H2O; 60 μm K2HPO4; 700 μm KH2PO4; 20 μm FeSO4, 7H2O; 20 μm Na2EDTA, 2H2O; 75 nm (NH4)Mo7O24, 4H2O; 3.5 μm MnSO4, H2O; 3 μm ZnSO4, 7H2O; 9.25 μm H3BO3; 785 nm CuSO4, 5 H2O; final pH 5.8] with additional 1% (w/v) Suc and 0.8% (w/v) bactoagar in the case of the solid medium. For metal tolerance tests, seeds were germinated on bactoagar nutrient solution in the presence of heavy metals [CdCl2, ZnSO4, CoCl2, and Pb(CH3COO)2].
EGFP∷AtHMA3, AtHMA3∷EGFP Fusions, and Expression
Full-length cDNA was obtained by RT-PCR on total RNA extracted from leaves of Arabidopsis (ecotype Ws) as described previously (Gravot et al., 2004). Gateway ends were added by PCR, and the construction was then subcloned in the plant Gateway vector pMDC43 (Curtis and Grossniklaus, 2003) by LR Clonase recombination (Invitrogen). Transgenic plants were generated by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain C58 and selected on hygromycin B (30 μg mL−1). For fluorescence confocal microscopy, 3-week-old plants were used.
Protoplasts of Arabidopsis mesophyll cells were isolated and transfected with the cDNAs of the EGFP∷AtHMA3 and the AtHMA3∷EGFP (this last construction was subcloned in the plant Gateway vector pMDC83 as described above) translational fusions or EGFP alone by the polyethylene glycol method described previously (Abel and Theologis, 1995; Kovtun et al., 2000). Typically, 0.1 mL of protoplast suspension (2 × 105 mL−1) was transfected with 30 to 50 μg of cDNA in the presence of 40% (w/v) polyethylene glycol for 30 min at 23°C. They were then washed with W5 solution (150 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, and 1.5 mm MES-KOH, pH 5.6) and incubated in W5 solution overnight at 17°C before observations. Transient expression experiments were repeated three times. Images were collected using a confocal laser scanning microscope (LC TCS-SP2; Leica) with a 488-nm argon laser for excitation. An objective HCX Plan Apo CS 63 oil (Leica) was used. The fluorescence signals were collected from 500 to 530 nm for EGFP.
GUS Activity
The region upstream of AtHMA3 from −2,747 to 0, in reference to the ATG start codon, +1, was amplified by PCR; the primers were used to add the Gateway ends. The fragment was then recombined to the pMDC162 vector, as described in the previous section. Generation and selection of transgenic plants were performed as described in the previous section. Plants or organs, at different stages of their development, were examined for GUS activity according to Jefferson et al. (1987). The samples were vacuum filtrated in the GUS staining solution, 50 mm NaPO4, pH 7.0, 0.01% (w/v) Triton X-100, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide and then incubated at 37°C for 4 h. Immediately after, pigments were washed by incubation of stained samples in 70% (v/v) ethanol at 60°C for 2 h. To perform transverse sections, plantlet roots and leaves were fixed in resin, after desiccation in successive ethanol baths of rising concentrations, for 30 min each.
Obtention of AtHMA3-Overexpressing Lines
The cDNAs of AtHMA3 and the translational fusion EGFP∷AtHMA3 were subcloned in a pGREEN0179 binary vector. These constructions were then introduced to AGL1 cells of A. tumefaciens by electroporation. The agrotransformation of Ws plants was carried out by the floral dip method; the transformant plants were selected on solid medium supplemented with hygromycin B (30 μg mL−1).
Metal Content Analysis
The germination of surface-sterilized seeds of wild-type Ws and 35S∷AtHMA3 lines was carried out on solid medium. After 2 weeks, the plantlets were placed on sand, left there for an additional 3-week period, and finally transferred to a hydroponic culture home-built setup. The nutrient solution and the toxic metal were replenished every 2 d; Cd was supplied at 30 μm. Hydroponically grown plants were harvested after 11 d of metal treatment. Roots were rinsed with 10 mm EDTA and then with distilled water. Roots and leaves were dried for 48 h at 50°C and mineralized. The metal content of these plants was determined using ICP-AES (Vista MPX; Varian).
Protoplast Cd Loading and Fluorescence Measurements
Leaf protoplasts from wild-type and 35S∷AtHMA3-overexpressing lines were isolated from 3-week-old seedlings by the enzymatic method as follows. The leaves were incubated overnight, at room temperature, in buffer A (0.6 m sorbitol, 5 mm MES/KOH, pH 5.5, 0.5 mm CaCl2, 0.5 mm MgCl2, 0.5 mm ascorbic acid, 0.25% [w/v] bovine serum albumin, 0.02% [w/v] Pectolyase Y-23, and 1.5% [w/v] Cellulase RS), then the protoplasts were filtered and collected by centrifugation (110g, 10 min). Next, the protoplast pellet was resuspended in buffer B (0.1 mm CaCl2, 10 mm KCl, 0.5 m sorbitol, 0.05% [w/v] polyvinylpyrrolidone 40, 0.2% [w/v] bovine serum albumin, and 5 mm MES, pH 5.5). A stock solution of BTC-5N, in the acetoxymethyl ester form (Molecular Probes), was prepared and used as described previously (Haugland, 1994). The protoplasts were incubated in the presence of BTC-5N for 50 min at 4°C and centrifuged, and the pellet was resuspended into 0.5 mL of buffer B. For detection of Cd in samples, the protoplasts were incubated in the presence of 0.35 mm Cd for 5 min in darkness. Fluorescence was visualized after excitation at 415 nm using a confocal laser scanning microscope. An objective HCX Plan Apo CS 63 oil (Leica) was used. The fluorescence signal was collected from 500 to 530 nm as described previously (Haugland, 1994). Laser intensity was set at about 40% to 45%, and the gain and offset adjustments were not modified between the samples. Fluorescence quantification was performed on eight to 12 protoplasts for each genotype and condition using ImageJ 1.36 software (http://rsb.info.nih.gov/ij/).
Genomic DNA Extraction and PCR Experiments
Genomic DNA was extracted from the inflorescence parts of various Arabidopsis ecotypes (Table I). For each extraction, two inflorescences were ground with a medium containing 100 mm Tris-HCl, pH 8.2, 50 mm EDTA, 100 mm NaCl, 0.1% (w/v) SDS, and 0.1 mg mL−1 proteinase K. The samples were incubated for 10 min at 37°C, and then 500 mL of phenol-chloroform-isoamyl alcohol was added. After 5 min at 37°C, the samples were centrifuged for 5 min at 14,000g at 4°C, and 50 mL of Na(CH3COO) (3 m), pH 5.2, and 500 mL of isopropanol were added. After centrifugation at 14,000g for 5 min, the pellets were resuspended in a solution of 500 mL of 1× Tris-EDTA buffer. The samples were incubated for 30 min at 37°C in the presence of 2 mL of RNase A (10 mg mL−1). Next, 500 mL of chloroform-phenol-chloroform-isoamyl alcohol was then added, and the samples were centrifuged for 5 min at 14,000g at 4°C. The supernatants were collected and precipitated with 50 mL of Na(CH3COO) (3 m), pH 5.2, and 500 mL of cold absolute ethanol. The DNA pellets were obtained after 15 min of centrifugation at 14,000g at 4°C. The pellets were washed with 70% (v/v) ethanol and resuspended in 1× Tris-EDTA buffer. Independent PCR experiments were carried out on these genomic DNAs with pfuUltra High-Fidelity DNA Polymerase (Stratagene) and a primer pair around the base pair deletion found in the AtHMA3 gene of the Col-0 ecotype. Different PCR products independently obtained from each genomic DNA ecotype were sequenced.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY055217.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignments of AtHMA2 and AtHMA3 according to ClustalW multiple alignment.
Supplemental Figure S2. AtHMA3 transcripts are enhanced in 35S∷AtHMA3 and 35S∷EGFP∷AtHMA3-overexpressing plants.
Supplemental Figure S3. Root lengths of wild-type and various 35S∷AtHMA3 and 35S∷EGFP∷AtHMA3-overexpressing lines under metal stress.
Supplemental Movie S1. Confocal imaging in z-axis of a Ws protoplast, loaded with BTC-5N, as described previously in the legend of Figure 5.
Supplemental Movie S2. Confocal imaging in z-axis of a Ws protoplast, loaded with BTC-5N after a 5-min 0.35 μm Cd treatment.
Supplemental Movie S3. Confocal imaging in z-axis of a 35S∷AtHMA3 protoplast, loaded with BTC-5N, as described previously in the legend of Figure 5.
Supplemental Movie S4. Confocal imaging in z-axis of a 35S∷AtHMA3 protoplast, loaded with BTC-5N, after a 5-min 0.35 μm Cd treatment.
Supplementary Material
Acknowledgments
We thank Serge Chiarenza for helpful technical assistance in the preparation of root and leaf cuts and GUS observations. We acknowledge Prof. Chris Cobbett and Dr. Narelle Cairns for the generous gift of Athma3-1 knockout seeds and Dr. Thierry Desnos for the generous gift of various Arabidopsis ecotype seeds. We are grateful to Michelle Turek for having carefully looked over the manuscript.
This work was supported by the Commissariat à l'Energie Atomique and the Toxicologie Nucléaire Environnementale program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pierre Richaud (pierre.richaud@cea.fr).
The online version of this article contains Web-only data.
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8 87–96 [DOI] [PubMed] [Google Scholar]
- Andrés-Colas N, Sancenon V, Rodriguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L (2005) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45 225–236 [DOI] [PubMed] [Google Scholar]
- Argüello JM (2003) Identification of ion-selectivity determinants in heavy-metal transport P1B-ATPases. J Membr Biol 195 93–108 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46 84–101 [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37 251–268 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212 475–486 [DOI] [PubMed] [Google Scholar]
- Clemens S (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88 1707–1719 [DOI] [PubMed] [Google Scholar]
- Clemens S, Palmgren MG, Krämer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7 309–315 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2006) Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol 9 322–330 [DOI] [PubMed] [Google Scholar]
- Curtis M, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren E, Argüello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P1B-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraustro da Silva JJR, Williams RJP (2001) The Biological Chemistry of the Elements, Ed 2. Oxford University Press, New York
- Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P (2004) AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett 561 22–28 [DOI] [PubMed] [Google Scholar]
- Hall JL (2002) Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot 53 1–11 [PubMed] [Google Scholar]
- Haugland RP (1994) Spectra of fluorescent dyes used in flow cytometry. Methods Cell Biol 42 Part B 641–663 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS (2007) A novel major facilitator superfamily protein at the tonoplast influences Zn tolerance and accumulation in Arabidopsis. Plant Physiol 143 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97 383–393 [DOI] [PubMed] [Google Scholar]
- Howden R, Andersen CR, Goldsbrough PB, Cobbett CS (1995. a) A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol 107 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R, Goldsbrough PB, Andersen CR, Cobbett CS (1995. b) Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol 107 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenkov V, Hirschi K, Crutchfield JD, Wagner GJ (2007. a) Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 226 1379–1387 [DOI] [PubMed] [Google Scholar]
- Korenkov V, Park S, Cheng NH, Sreevidya C, Lachmansingh J, Morris J, Hirschi K, Wagner GJ (2007. b) Enhanced Cd2+-selective root-tonoplast-transport in tobaccos expressing Arabidopsis cation exchangers. Planta 225 403–411 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer U, Talke IN, Hanikenne M (2007) Transition metal transport. FEBS Lett 581 2263–2272 [DOI] [PubMed] [Google Scholar]
- Küpper H, Seib LO, Sivaguru M, Hoekenga OA, Kochian LV (2007) A method for cellular localization of gene expression via quantitative in situ hybridization in plants. Plant J 50 159–175 [DOI] [PubMed] [Google Scholar]
- Lebrun M, Audurier A, Cossart P (1994) Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol 176 3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA (1996) The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem 271 6509–6517 [DOI] [PubMed] [Google Scholar]
- Lindberg S, Landberg T, Greger M (2004) A new method to detect cadmium uptake in protoplasts. Planta 219 526–532 [DOI] [PubMed] [Google Scholar]
- Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97 4956–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J 32 539–548 [DOI] [PubMed] [Google Scholar]
- Puig S, Mira H, Dorcey E, Sancenón V, Andrés-Colás N, Garcia-Molina A, Burkhead JL, Gogolin KA, Abdel-Ghany SE, Thiele DJ, et al (2007) Higher plants possess two different types of ATX1-like copper chaperones. Biochem Biophys Res Commun 354 385–390 [DOI] [PubMed] [Google Scholar]
- Rensing C, Mitra B, Rosen BP (1997) The zntA gene of Escherichia coli encodes a Zn (II)-translocating P-type ATPase. Proc Natl Acad Sci USA 94 14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Cavet JS, Robinson NJ (1999) Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J Biol Chem 274 25827–25832 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, Finazzi G, Grunwald D, Rappaport F, Vavasseur A, Joyard J, et al (2006) HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J Biol Chem 281 2882–2892 [DOI] [PubMed] [Google Scholar]
- Sharma R, Rensing C, Rosen BP, Mitra B (2000) The ATP hydrolytic activity of purified ZntA, a Pb (II)/Cd (II)/Zn (II)-translocating ATPase from Escherichia coli. J Biol Chem 275 3873–3878 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Song WY, Sohn EJ, Martinoia E, Lee YJ, Yang YY, Jasinski M, Forestier C, Hwang I, Lee Y (2003) Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol 21 914–919 [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Krämer U (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576 306–312 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Prévéral S, Forestier C, Vavasseur A, Richaud P (2005) Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett 579 1515–1522 [DOI] [PubMed] [Google Scholar]
- Williams LE, Mills RF (2005) P(1B)-ATPases: an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10 491–502 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Kieber JL (2000) A strong loss-of-function mutation RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell 12 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.