Abstract
Endoreduplication, a modified cell cycle that allows cells to increase ploidy without subsequent cell division, is a key component of plant growth and development. In this work, we show that some, but not all, of the endoreduplication of Arabidopsis (Arabidopsis thaliana) is mediated by the expression of a WD40 gene, FIZZY-RELATED2 (FZR2). Loss-of-function alleles show reduced endoreduplication and reduced expansion in trichomes and other leaf cells. Misexpression of FZR2 is sufficient to drive ectopic or extra endoreduplication in leaves, roots, and flowers, leading to alteration of cell sizes and, sometimes, organ size and shape. Our data, which suggest that reduced cell size can be compensated by increased cell proliferation to allow normal leaf morphology, are discussed with respect to the so-called compensation mechanism of plant development.
A key element of plant morphogenesis is the balance among cell proliferation, expansion, and differentiation to produce organs of the characteristic sizes and shapes. At the heart of plant morphogenesis is the cell versus organism debate. On one hand, the cell theory postulates that the cells of a multicellular organism behave autonomously, and the sum of their activities results in the morphology of that organism (Schwann, 1839). On the other hand, the organismal theory postulates that morphology is governed by a genetic mechanism separate from the behavior of individual cells (Kaplan and Hagemann, 1991). Regardless of perspective, it is clear that the plant program of cell growth and proliferation is dynamic and responsive, i.e. perturbations in one factor can be compensated by modifications in the other (Day and Lawrence, 2000; Mizukami, 2001). For example, forcing a decrease in leaf cell number by overexpressing the cell cycle regulator KRP2 results in an increase in cell volume (De Veylder et al., 2001). Conversely, an increase in cell volume caused by the overexpression of ABP1 leads to a decrease in cell number (Jones et al., 1998). This compensation mechanism has been incorporated into a hybrid cell/organismal theory called “neo-cell theory” (Tsukaya, 2003). The nature of this compensation mechanism is unclear, although it likely involves cellular reaction to organ-level positional information.
During the development of many plants, certain cell types undergo extensive endoreduplication, a modified cell cycle that results in DNA replication without subsequent mitosis. In leaves of Arabidopsis (Arabidopsis thaliana), several rounds of endoreduplication occur in trichomes and in most epidermal pavement cells, while guard cells remain diploid (Melaragno et al., 1993). In the last decade, extensive research has looked at the trichome as a model to unravel the mechanism controlling endoreduplication. During trichome development, four rounds of endoreduplication regularly take place, although variation exists (Melaragno et al., 1993). Some Arabidopsis mutants show concomitant changes in trichome ploidy and branch number, supporting a role of endoreduplication in the control of branch number (Hülskamp et al., 1994; Perazza et al., 1999; Downes et al., 2003). Branching changes are not always tied to endoploidy changes, however; for example, plants with loss-of-function alleles or overexpression of STICHEL show trichome branch number alterations without changes in nuclear DNA content (Ilgenfritz et al., 2003). Endoreduplication appears to be an important determinant of cell and organ size. Indeed, a correlation between cell size and endoploidy has been found in Arabidopsis and other plant species (Galbraith et al., 1991; Traas et al., 1998; Kudo and Kimura, 2002), although Arabidopsis root cells constitute an exception (Beemster et al., 2002), and certain genetic manipulations can also uncouple this relationship (Hemerly et al., 1993; Wang, et al., 2000; De Veylder et al., 2001; Fujikura et al., 2007). Interestingly, different levels of endoreduplication may affect the same tissue in distinct species. For instance, orchid (Oncidium varicosum and Phalaenopsis spp.) and cabbage (Brassica capitata) petals are large and undergo extensive endoreduplication, with some cells reaching 64C (Kudo and Kimura, 2001; Lee et al., 2004). The sizes of petal cells, and hence of petal organs, directly correlate with their ploidy in these studies. By contrast, the cone cells of Arabidopsis petals remain diploid despite being capable of endoreduplication (Hase et al., 2005), and this plant shows relatively small flowers in comparison to close relatives. Unlike orchid and cabbage, Arabidopsis is self-fertilizing, so producing small petals would not hinder its reproduction.
Similar cellular components regulate endoreduplication and the mitotic cell cycle. Cyclin-dependent kinases (CDKs) control the cell cycle by forming complexes with cyclins, by phosphorylation or dephosphorylation, or by association with CDK inhibitors. Expression and degradation of specific cyclins promote progression through distinct stages of the cell cycle and control the exit from the cell cycle. For example, D-type cyclins become phosphorylated and associate with the A-type CDK to induce the switch from G1 phase to S phase (Inzé and De Veylder, 2006). Destruction of B1-type cyclins seems to be necessary for completion of the M phase, because introduction of a B1 cyclin for which the destruction-box motif has been altered can prevent cytokinesis (Weingartner et al., 2004). In yeast and Drosophila, Fizzy-Related (FZR) family proteins have been shown to trigger the degradation of A- and B-type cyclins by targeting them to the Anaphase Promoting Complex/Cyclosome (APC/C), an E3 ubiquitin protein ligase (Sigrist and Lehner, 1997; Yamaguchi et al., 1997). In those studies, misexpression of FZR family proteins was able to induce endoreduplication. Furthermore, a recent study linked the APC/C to local cell expansion, endoreduplication, and the compensation mechanism in Arabidopsis (Serralbo et al., 2006).
The plant homologs of FZR were first implicated in endoreduplication in root nodules of Medicago sativa, and antisense expression of the Medicago truncatula Cell-cycle Switch 52 A and B (Ccs52A and Ccs52B), orthologs of FZR, led to reduced endoreduplication in that species (Cebolla et al., 1999). In Arabidopsis, three FZR homologs exist, and in cell cultures, FZR1 and FZR2 showed similar high expression in G1 and S phases of the cell cycle, while FZR3 expression increased at the end of G2 and beginning of M. All three FZR proteins associated to free and CDK-bound A- and B-type cyclins (Fülöp et al., 2005). Although the FZR genes are known to be expressed in all major tissues (Beemster et al., 2005), the normal developmental function of the FZR family has not yet been determined in Arabidopsis.
In this study, we examined the developmental roles of one of the Arabidopsis FZR homologs, FZR2. We found that FZR2 expression is necessary for correct cell expansion and endoreduplication, and its misexpression is sufficient to induce extra or ectopic endoreduplication and cell expansion. We gathered empirical data that address the putative compensation mechanism balancing cell proliferation with cell expansion.
RESULTS
FZR Genes Are Expressed Ubiquitously in Arabidopsis
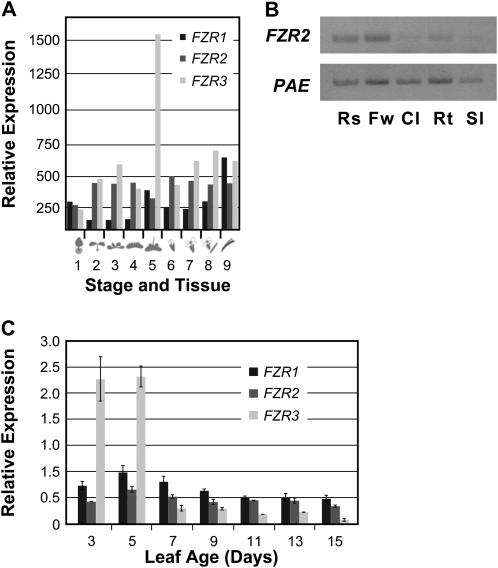
In our analysis of FZR function, we investigated the expression of these genes using Genevestigator (www.genevestigator.ethz.ch/). This revealed that they are relatively highly expressed in all organs throughout the life cycle (Fig. 1A). Other Genevestigator data indicated that FZR2 tends to be up-regulated when the plants are treated with brassinolide or cytokinins and down-regulated by treatment with abscisic acid or auxin. We confirmed FZR2 expression in seedlings, flowers, leaves, and roots by semiquantitative reverse transcription (RT)-PCR (Fig. 1B). We also assayed expression of FZR1, FZR2, and FZR3 during leaf development by quantitative real-time RT-PCR (qRT-PCR; Fig. 1C), and these data show relatively steady expression of FZR1 and FZR2 after peaks of expression at 2 to 5 d. Also in agreement with the Genevestigator summary, FZR3 was highly expressed in 3- and 5-d-old leaves. Subsequently, FZR3 expression dropped below relative levels of FZR1 and FZR2 expression.
Figure 1.
Expression of FZR genes. A, GENEVESTIGATOR developmental summary of FZR expression data from microarrays. Tissues examined are: (1) germinated seed, (2) seedling, (3) young rosette, (4) developed rosette, (5) bolting plants, (6) young flower, (7) developed flower, (8) flowers and siliques, and (9) mature siliques. B, RT-PCR data from Arabidopsis tissues: Rs, rosette leaves; Fw, flowers; Cl, cauline leaves; Rt, roots; Sl, seedlings. C, qRT-PCR of FZR1, FZR2, and FZR3, normalized against UBIQUITIN10. Error bars indicate sd.
fzr2-1 Leaf Cells Undergo Fewer Endoreduplication Cycles and Attain Smaller Sizes
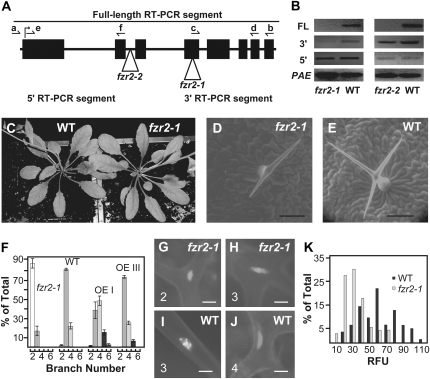
To investigate the function of FZR genes in plant development, we obtained two T-DNA insertion mutants for FZR2 from the SALK collection. The fzr2-1 (SALK_083656) and fzr2-2 (SALK_101689) lines carry insertions in the fourth exon and the second intron, respectively (Fig. 2A). Insertions were confirmed by sequencing. RT-PCR analysis showed that both alleles of fzr2 fail to produce full-length transcript, although mRNA segments upstream and downstream to the insertion were still detected (Fig. 2B). In both homozygous mutants, soil growth behavior and general morphology appeared wild type (Fig. 2C; data not shown). However, both homozygous mutants produced a similar trichome phenotype, i.e. a general reduction in trichome branch number (Fig. 2, D–F). Wild-type Col-0 leaves exhibit a prevalence of 3-branch and 4-branch trichomes, whereas the mutant leaves never show 4-branch trichomes but show mostly 2-branch trichomes (Fig. 2, D–F; data not shown). For both fzr2-1 and wild type, larger nuclei were associated with higher trichome branch numbers (Fig. 2, G–J). Both fzr2-1 and fzr2-2 displayed reductions in trichome branch number, and fzr2-1 was used for further analysis due to its exon-bound T-DNA. By in situ quantification of nuclear content, we determined that fzr2-1 leaves produce trichomes with a general 2-fold reduction in ploidy compared to wild type (Fig. 2K). From these data, we conclude that fzr2-1 trichome nuclei undergo fewer rounds of endoreduplication than wild type.
Figure 2.
Expression and phenotypes of loss-of-function fzr2 mutations. A, Diagram of the FZR2 locus depicting exons (black boxes), the locations of the two T-DNA insertion mutations (white triangles), and the locations of primer sequences used in the RT-PCR analyses shown in B (arrows). B, RT-PCR analysis of mutants and FZR2 OE representatives. The FL bands resulted from RT-PCR using primers a and b, the 3′ segment used primers c and d, and the 5′ segment used primers e and f. C, Flowering wild-type (WT; left) and fzr2-1 (right) plants. D and E, ESEM micrographs of leaf trichomes of fzr2-1 (D) and wild-type (E) plants. Scale bar = 100 μm. F, Summary of leaf trichome branch production. Error bars represent ses. G and J, Representatives of DAPI-stained trichome nuclei of fzr2-1 (G and H) and wild type (I and J). The branch number for each trichome is given in the lower left of each picture. Scale bar = 15 μm. K, Summary of in situ fluorescence measurements of DAPI-stained trichome nuclei, given in relative fluorescence units (RFU).
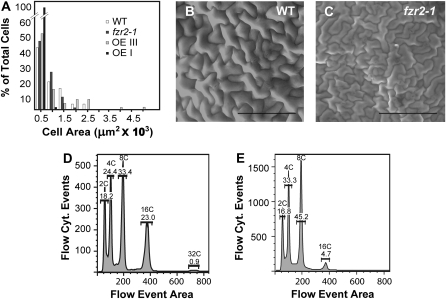
We examined leaf cells to further investigate the effect of fzr2 mutations on cell size and ploidy level. A comparison of the adaxial external surface areas of wild type and fzr2-1 epidermal pavement cells showed a statistically significant decrease in fzr2-1 cell size relative to wild type (Student's t test: P < 0.001; Fig. 3, A–C). Furthermore, flow cytometry of propidium iodide (PI)-stained nuclei released from whole leaf tissue revealed a general decrease in fzr2-1 endoreduplication compared to wild-type flow cytometry profiles (Fig. 3, D and E). The fzr2-1 leaves showed a much smaller percentage of 16C nuclei, suggesting that non-trichome leaf cells are also less able to undergo this later round of endoreduplication when FZR2 expression is disrupted. Taken together, the data are consistent with the reduction in cell size and ploidy seen in the trichomes.
Figure 3.
Comparisons of cell area and endoreduplication profiles of wild type and fzr2-1. A, Histogram of epidermal pavement cell surface areas of wild-type (WT; average [ave.] = 23,570 μm2; sd = 18,560 μm2), fzr2-1 (ave. = 16,250 μm2; sd = 12,820 μm2), OE I (ave. = 153.5 μm2; sd = 109.6 μm2), and OE III (ave. = 22,480 μm2; sd = 13,935 μm2) plants. Cell surface areas were binned into 500-μm2 categories. B and C, ESEMs of adaxial leaf surfaces of wild type (B) and fzr2-1 (C). Scale bar = 100 μm. D and E, Flow cytometric (cyt.) profiles of PI-stained leaf nuclei of wild type (D) and fzr2-1 (E). A total of 10,000 events were recorded for each run. Ploidy and percentage of total nuclei are given at the top of each peak.
Misexpression of FZR2 Is Sufficient to Drive Endoreduplication
While FZR2 function is necessary for endoreduplication in some leaf cells, it was unclear whether its expression is sufficient to drive endoreduplication. Expression of the FZR2 cDNA with the constitutive cauliflower mosaic virus 35S promoter (denoted OE) resulted in three phenotypic classes (Fig. 4A), each showing increased FZR2 transcript levels in developing leaves by qRT-PCR (Fig. 4B). Although development of these transgenic plants appeared normal during the first few days after germination, they produced trichomes with supernumerary branches and enlarged nuclei, and the size of a trichome nucleus was positively associated with its branch number (Fig. 4, C–G). Class I (OE I) was comprised of dwarfed, anthocyanin-pigmented plants showing small leaf cells (Figs. 3A and 4, A and H) and heavily over-branched trichomes, some producing as many 10 branches, although the majority produced three or four branches (Fig. 2F). Class II (OE II) plants were slightly larger and greener than OE I and showed fewer supernumerary branches. OE I and II plants arrested growth at 14 to 28 d after germination and almost always failed to bolt, while Class III (OE III) plants bolted and set seed as normal. OE III plants approached wild-type size but showed some very large cells (Figs. 3A and 4I), including trichomes that produced up to five branches (Fig. 2F), as well as many smaller cells. The aboveground tissue of OE III plants appeared normal at the macroscopic level (Fig. 4A). All three OE classes also showed an increase in the size of some guard cells, with about 16% readily identifiable as larger (sd 9%, n = 10 leaves; Fig. 5, C–F).
Figure 4.
Phenotypes associated with FZR2 OE lines. A, Wild type (WT; right) and OE classes 28 d past germination. B, Relative expression of wild type and the three OE classes by qRT-PCR, normalized to UBIQUITIN10 expression. Error bars represent sds. C, ESEM micrographs of a leaf trichome of class OE I showing six branches. Scale bar = 100 μm. D to G, Representatives of DAPI-stained trichome nuclei of OE I trichomes. The branch number is given in the lower left of each picture. Scale bar = 15 μm. H and I, ESEMs of adaxial leaf surfaces of OE I (H) and OE III (I). Scale bar = 100 μm.
Figure 5.
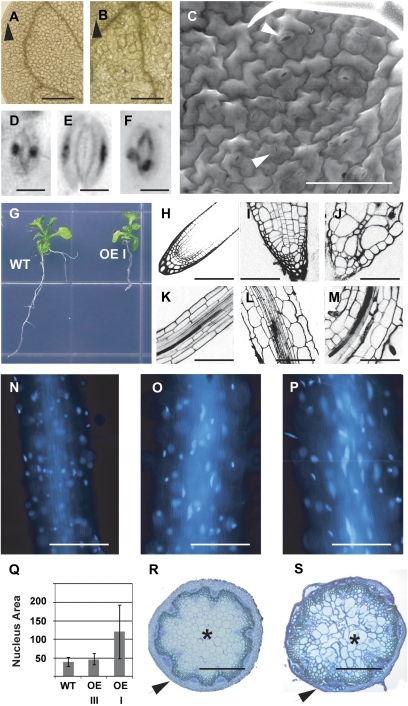
Cotyledon, root, stem, and guard-cell phenotypes of FZR2 OE lines. A and B, Cleared cotyledon of wild type (A) showing smaller mesophyll cells than those of an OE line (B). Black arrowheads indicate normal-sized mesophyll cells. Scale bar = 500 μm. C, ESEM of the adaxial surface of an OE I plant, showing both normal-sized (lower arrowhead) and enlarged (upper arrowhead) stomata. Scale bar = 50 μm. D to F, Light micrographs of DAPI-stained stomata of wild-type (D) and OE I plants (E and F). Scale bar = 10 μm. G, Root growth of wild-type (WT) and OE Class I seedlings after 9 d of growth on vertical 1.5% agar plates. H to M, Confocal images of PI-stained root tips of wild type (H), OE III (I), and OE I (J), and root mature zones of wild type (K), OE III (L), and OE I (M). Scale bar = 100 μm. N to P, DAPI-stained nuclei of root mature zones of wild type (N), OE III (O), and OE I (P). Q, Measurements of nuclear area of root cortex cells of wild-type and OE roots. Wild-type root cortex cell nuclei averaged 3,972 ± 1,179 μm2 (n = 46), OE III nuclei averaged 4,721 ± 1,529 μm2 (n = 22), and OE I nuclei averaged 12,090 ± 7,180 μm2 (n = 24). Student's t test (two-tailed with unequal variance) showed OE III to be marginally significantly different from wild type (P = 0.043), while OE I was very significantly different (P = 5.2 × 10−6). The OE I and OE III nuclei were also very significantly different from each other (P = 2.2 × 10−6). R and S, Cross sections of stems of wild-type (R) and OE III (S) plants. Black arrowheads indicate cortex tissue and black asterisks indicate pith tissue. Scale bar = 10 μm.
Root development was dramatically altered in the OE plants (Fig. 5G), although the severity of defects varied among the phenotypic classes. OE I plants showed the most severe deformations in root architecture, being thicker and composed of larger cells than wild type (Fig. 5, J and M). The overall shape of the OE I root was still cylindrical, but the characteristic cell layers were not fully conserved. In comparison, OE II and OE III roots were moderately and mildly disrupted, respectively, with less extreme increases in root width and cell sizes (Fig. 5, I and L). The increases in root cell sizes correlated with increases in nuclear sizes (Fig. 5, N–Q). The nuclear size increases were statistically significant (P < 0.05) when comparing either OE I or OE III to wild type using a Student's t test (Fig. 5Q). Another noteworthy effect of 35S∷FZR2 expression was a moderate increase in inflorescence stem thickness in all three OE lines associated with an increase in vascular tissue cell size. This was accompanied by a shift in tissue proportions, such that the epidermal layer appeared to be thinner (Fig. 5, R and S).
Approximately 3% of T2 generation OE transgenic lines (two of 66 independent T0 lines) showed incomplete transgene silencing that prevented FZR2 overexpression early yet apparently allowed the transgene to escape silencing effects later in plant development. This resulted in what appeared to be mosaic sectors of over-endoreduplicating cells, such that regions of enlarged cells and over-branched trichomes were detected within leaves that were otherwise composed of normal-sized cells and 3- or 4-branch trichomes. As increased trichome branching often indicates increased trichome endoreduplication, it is likely that the trichomes and the other cells in the sector had overexpressed FZR2. Subsequent generations of these lines failed to exhibit this phenomenon in postembryonic tissues but produced sectors in the cotyledons. An example of this is shown in Figure 5B, in which the cotyledon mesophyll cells of a semi-silenced 35S:FZR2 line are greatly enlarged in comparison to wild-type cotyledon mesophyll cells (Fig. 5A). The sizes of epidermal cells and of the entire organ did not differ between wild-type and transgenic lines.
FZR2 Expression Is Sufficient to Drive Endoreduplication in Diploid Petal Tissue
The 35S∷FZR2 misexpression experiments showed that more FZR2 activity led to extra endoreduplication. However, this endoreduplication resulted in tissue defects that hindered the plant's general growth, resulting in dwarfism in the lines that misexpressed FZR2 highly (i.e. OE I and II). For example, a loss of root tissue function is likely to result from the tissue disorganization seen in OE I roots (Fig. 5, J and M), leading to secondary defects due to the perturbation of nutrient and water uptake for the rest of the plant. This complicated the interpretation of FZR2 function in plant development. To circumvent these types of secondary defects, we used a tissue-specific promoter to limit the extra function. We also wished to address whether FZR2 misexpression could drive endoreduplication in cells that normally lack it entirely. The cone cells of Arabidopsis petals do not undergo endoreduplication (Hase et al., 2005) and show nearly uniform shape and size in fully developed petal tissue (Fig. 6G). Therefore, we created transgenic plants expressing FZR2 in cells of the floral primordia that generate B-whorl organs, using the APETELA3 (AP3) promoter. The AP3 promoter drives expression in flower buds shortly after the sepals emerge at stage 3 in a zone of cells that will give rise to the petal and stamen primordia (Smyth et al., 1990; Jack et al., 1992). Expression continues in the petals and stamens as they develop until the time of fertilization (Jack et al., 1992; Hill et al., 1998).
Figure 6.
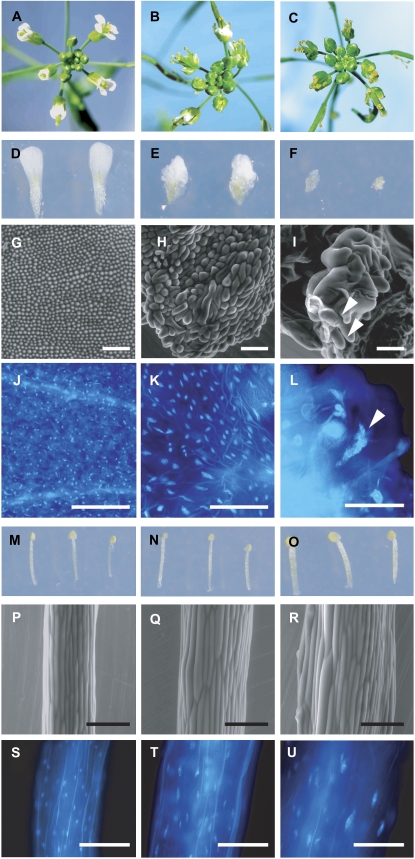
Phenotypes associated with wild-type and AP3∷FZR2 flowers. A, D, G, J, M, P, and S show floral tissue from wild type; B, E, H, K, N, Q, and T show AP3∷FZR2 weak phenotypes; C, F, I, L, O, R, and U show AP3∷FZR2 strong phenotypes. A to C, Inflorescences. D to F, Dissected petals. G to I, ESEM micrographs of petal epidermal cells. In I, white arrowheads indicate the multiple conical peaks of the giant cells. J to L, UV fluorescence micrographs of DAPI-stained petals. In L, a white arrowhead indicates a giant nucleus. M to O, Dissected stamens. P to R, ESEM micrographs of stamen filament. S to U, UV fluorescence micrographs of DAPI-stained stamens. Scale bar = 100 μm.
AP3∷FZR2 lines showed an increase in cell size in both stamens and petals, and this cell-size increase was matched by nuclear size increase (Fig. 6). Compared with wild-type petal nuclei, the AP3∷FZR2 petal nuclei were extremely enlarged (Fig. 6, J–L). The petals nonetheless retained petal differentiation characteristics, despite the severe deviation from their normal developmental program. For instance, wild-type petal cone cells show ridges along the cone-shaped surface (Supplemental Fig. S1A). In the giant AP3∷FZR2 petal cells, several conical peaks were seen instead of the single peak seen for a wild-type cell (Fig. 6I), and the characteristic ridges were present on each (Supplemental Fig. S1B). The petals lost their flat shape, and instead the tissue grew outward. AP3∷FZR2 stamens were much wider with larger nuclei (Fig. 6, S–U) and showed more and larger epidermal cells, although their length and general shape were similar to wild type (Fig. 6, M–R).
Another interesting phenotype involved the shapes of the nuclei themselves. Previous work suggests that increased nuclear content often corresponds with increased spatial separation of chromatin within plant nuclei (Kato and Lam, 2003). Indeed, nuclei seemed to keep roughly spherical shapes and uniform DNA distribution at lower ploidies when examined using 4′-6-diamidino-2-phenylindole (DAPI) and epifluorescence microscopy (e.g. wild-type root nuclei; Fig. 5N). At higher ploidies, however, we observed nuclei in ellipsoid and spindle shapes (Fig. 5, O and P). We further observed that the larger OE trichomes possessed nuclei that seemed to have spread out even more (Fig. 4, D–G). Brighter DAPI-stained DNA was seen in punctate bunches at various locations, possibly at heterochromatic regions (e.g. Fig. 4G). Moreover, a positive relationship was noted among trichome branch number, nuclear size, and degree of chromatin separation (Fig. 4, D–G).
DISCUSSION
FZR2 Expression Is Necessary and Sufficient for Some Endoreduplication in Arabidopsis
This article demonstrates that loss of FZR2 function in the fzr2 mutants led to reduced endoreduplication in trichomes along with decreased trichome branching. Although truncated FZR2 mRNA fragments remain and could produce truncated proteins, we do not expect this to contribute to the phenotypes, because heterozygotes appear wild type. Similar correlations between branching and ploidy have been observed in other 2-branch trichome mutants (Hülskamp et al., 1994; Perazza et al., 1999). Likewise, there was an overall decrease in the ploidy levels of fzr2-1 leaf cells. Flow cytometric analysis of fzr2-1 leaves indicated that the mutants were unhindered in undergoing up to two endocycles in this tissue, but endoreduplication past the 8C level was greatly reduced. As such, we conclude that FZR2 is a factor controlling the induction of early rounds of endoreduplication. The remaining rounds of endoreduplication may be mediated by the other two FZR family homologs, FZR1 and FZR3, because all of these genes are expressed in the same tissues in leaves, although to different degrees (Fig. 1, A–C). It should, however, be cautioned that FZR3 probably differs in time of expression during the cell cycle (Tarayre et al., 2004; Fülöp et al., 2005). Furthermore, FZR2 may have other functions, separate from its role in endoreduplication, because it is expressed in nonendocycling tissues as well (Beemster et al., 2005). That FZR2 can induce endoreduplication is also supported by the observation that its misexpression caused an increase in nuclear and cell sizes in both root and leaf tissues and promoted endoreduplication and cell expansion in petal tissues that do not normally undergo these processes.
Cell Sizes, Organ Sizes, and the Compensation Mechanism
Despite the absence of most 16C and 32C cells, the leaves in the loss-of-function mutants were indistinguishable from wild type at the organ level. Because the leaves lacked most of the large 16C cells, they must have consisted of smaller but more numerous cells; this was evident in the external surface area measurements of the epidermal cells (Fig. 3A). As the cells were deficient in the 8C to 16C endocycle, the resulting space must have been filled by proliferation of the 2C and 4C cells. It was unclear whether this was due to extra proliferation and endoreduplication from 2C cells or whether the 4C and 8C cells were also capable of dividing. The volume was not accounted for by extra 2C cells alone, however, because the 4C and 8C cells increased in proportion in our flow cytometric analyses, while the 2C cells did not (Fig. 3, D and E). Endoreduplication is generally thought to preclude subsequent cell division (Sugimoto-Shirasu and Roberts, 2003), although there is evidence that endoreduplicated trichome socket cells can reenter mitosis under certain conditions (Weinl et al., 2005). If endoreduplicated leaf cells were becoming mitotically active in response to a reduced size, we might expect the trichomes to have been able to reach a wild-type size and shape but as a multicellular form, as documented previously in the siamese mutant trichomes (Walker et al., 2000). Given the absence of multicellular trichomes in fzr2 mutants, we favor an increase in cell number due to proliferation by 2C cells, followed by further endoreduplication to 4C and sometimes 8C.
The cause of the extra proliferation is not clear, but several explanations should be considered. First, it could be that FZR2 normally functions early in leaf growth to switch cells from proliferation to endoreduplication. If this were the case, the early endocycles of fzr2-1 could instead be mitotic cycles, and perhaps FZR1 or FZR3 would trigger the later endocycles. From the cell theory perspective, the cells might have a certain number of total cycles (or developmental window of cycling), and the shift toward mitotic cycles would be at the expense of later endocycles. Contrary to this hypothesis, however, is the lack of trichome subdivision despite a reduction of endoreduplication. Second, FZR2 could be a factor driving later endocycles. Arabidopsis leaf cells proliferate and then endocycle along a basipetal gradient (Donnelly et al., 1999; Beemster et al., 2005). In fzr2-1 leaves, FZR1 or FZR3 might induce the switch from proliferation to expansion as normal. During the expansion phase, however, some cells would be unable to reach the appropriate sizes and ploidies due to lack of FZR2. It is possible that the defect could be sensed through positional cues, and a proliferation signal could be transmitted from the expanding cells to cells with the potential for further mitosis. This organismal-theory hypothesis would explain the remarkable result of normal leaf morphogenesis despite the restriction of cell growth. Third, it is possible that the overproliferation in fzr2-1 results from accelerated proliferation in a manner similar to a recent 35S:CYCD2;1 report (Qi and John, 2007), in which overabundant CycD2;1 led to precocious cell divisions and smaller, more numerous cells. The 35S:CYCD2;1 leaves were also of normal morphology, but leaf nuclei were not evaluated for endoreduplication, nor were the trichomes abnormal (Qi and John, 2007). Unlike A- and B-type cyclins, D-type cyclins do not contain the D-box motif that putatively facilitates their proteolysis, so we do not expect CYCD2;1 overabundance and accelerated cell divisions in fzr2-1. Furthermore, expression in tobacco (Nicotiana tabacum) of a nondegradable cyclin B resulted in cell shape defects and phragmoplast failure instead of mitotic acceleration (Weingartner et al., 2004). Finally, a study using 35S:ANT transgenic Arabidopsis showed that cell overproliferation does not necessarily lead to reduced cell size (Mizukami and Fischer, 2000).
Hence, while none of the hypotheses can be eliminated at this point, the first two seem more likely. Differentiating between them will be difficult, unfortunately, because fzr2-1 growth kinematics would be the same for both. Nevertheless, this study may be the first example of organ developmental compensation via increased cell proliferation to attain a normal leaf size. Decreases in leaf cell volume have been observed in other studies, but in such cases, normal organ size was not attained. One such study showed that rot3 leaves had the same number of cells as wild type, yet these cells were shorter along the proximodistal axis and hence the leaves were shorter (Tsuge et al., 1996). Similarly, overexpressing a homeobox gene (AtHB13) or expressing an antisense sequence of an expansin gene (AtEXP10) caused a reduction in cell size with no compensatory increase in cell number (Cho and Cosgrove, 2000; Hanson et al., 2001). Several other studies have proposed that when cell number is reduced, cell volume increases to compensate, or that when cell volume is increased, cell proliferation decreases (Jones et al., 1998; Wang et al., 2000; De Veylder et al., 2001; Bemis and Torii, 2007). In the same manner, FZR2 OE III plants showed an increase in leaf cell size and a decrease in cell number, such that the wild-type and transgenic leaves were similar in shape and size despite the presence of fewer and larger cells (although the smallest cells of OE III in Fig. 3A were more numerous, perhaps because they were restricted from expanding by the larger cells). Leaves of the transgenic plants showing phenotype OE I and OE II, however, lacked this compensation effect, showing smaller organs composed of smaller cells. One explanation for this could be that the OE I and OE II leaf cells were unable to enter S phase due to the misexpression of FZR2, and hence neither endoreduplication nor proliferation sufficed, while the milder overabundance in OE III decreased cell proliferation but allowed extra endoreduplication. Similar concentration-dependent effects were seen for overexpression of ICK/KRP1 (Weinl et al., 2005). An alternative, less direct explanation may relate to the severe physiological defects that may result from FZR2 overexpression. Indeed, the severity of above-ground phenotypes correlated with the degree of root architecture distortion in FZR2 overexpressing plants (Figs. 4A and 5, H–M). Therefore, it is possible that the dwarf phenotype of OEI and OEII plants may have been caused by general nutrient deficiency and dehydration. In agreement with this explanation, OE I and OE II plants accumulated anthocyanins, a phenotype often associated with nutrient stress (Steyn et al., 2002). These two explanations for the dwarf phenotype of OE I and OE II plants postulate that the expression levels of the 35S∷FZR2 transgene were commensurate with the severity of defects in roots and above-ground organs. Indeed, the steady-state levels of FZR2 transcript were higher in OE I and OE II plants relative to OE III (Fig. 4, A and B).
Similar to the leaf overexpression effects in OE III, we saw a decrease in cell number and an increase in cell size in the petals of AP3∷FZR2 plants, although here, the organs themselves were much smaller (Fig. 6). Normally lacking endoreduplication, the petal cells were clearly competent to undergo extensive endocycles upon FZR2 misexpression. The guard-cell size increase was yet another indication that misexpression of FZR2 can induce ectopic endoreduplication (Fig. 5, C–F).
Petal Cells Can Endoreduplicate Yet Retain Petal Cell Characteristics
The portion of the AP3 promoter used to drive FZR2 expression in petals and stamens has been shown to direct expression in the floral meristem prior to the appearance of both petal and stamen primordia (Jack et al., 1992; Hill et al., 1998). Ectopic endoreduplication early in AP3∷FZR2 petal development likely triggered the cells to exit the proliferation phase and enter the endocycle, preventing normal petal cell proliferation. Plants with the highest AP3∷FZR2 expression at that early phase of petal development would form flowers with the largest petal cells and nuclei at the end of development (Fig. 6, F and I; Supplemental Fig. S1, A and B). The presence of giant cells and nuclei in some AP3∷FZR2 lines suggest that these cells underwent high numbers of endoreduplication cycles. Similar extreme tissue disorganization phenotypes were also found in plants with other alterations in the cell cycle, such as expression of non-degradable cyclin B or plants engineered to lose the HOBBIT CDC27 ortholog postembryonically (Weingartner et al., 2004; Serralbo et al., 2006). It is remarkable that these giant petal cells retain petal cone cell characteristics (e.g. ridges and conical shapes; Supplemental Fig. S1B) despite the severe perturbation to their developmental program. More normal organ morphology was seen for transgenic stamens, although there was an increase in both cell size and number.
Our results show that with the loss of FZR2 function, endoreduplication cycles past 8C are infrequent. In petals, expression of FZR2 led to de novo endoreduplication, yet the enlarged cone cells retained tissue characteristics. In conclusion, we have shown that FZR2 is a regulator of cell growth and endoreduplication, and we have provided data that support the presence of a morphological compensation mechanism that allows plant organs to compensate for reduced cell size by increasing proliferation.
MATERIALS AND METHODS
Gene Nomenclature
The FZR gene family includes FZR1 (At4g11920), FZR2 (At4g22910), and FZR3 (At5g13840). We refer to these genes using the FZR three-letter symbol as per the conventions set out by the Arabidopsis (Arabidopsis thaliana) research community (www.arabidopsis.org) to replace the previously used nonstandard symbols for these genes, as follows: FZR1 (CCS52A2), FZR2 (CCS52A1), and FZR3 (CCS52B; Cebolla et al., 1999).
Plant Material and Growth Conditions
All Arabidopsis alleles were derived from the Columbia ecotype. Two T-DNA insertions into the At4g22910 locus (FZR2) were acquired from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH): SALK_083656 (fzr2-1) and SALK_101689 (fzr2-2). Insertion lines were back-crossed into the Columbia-0 accession three times to eliminate secondary mutations. Primers used for genotyping are listed in Supplemental Table S1. In vitro plant culture was performed on half-strength Murashige and Skoog medium (MP Biomedicals), supplemented with 0.9% Suc and 1% agar, and adjusted to pH 5.8. Soil-grown plants were raised on a 2:1 ratio of Sunshine coarse vermiculite to Sunshine potting mix (SunGro Horticulture). First watering was supplemented with 1 teaspoon of Jack's Classic 20-20-20 fertilizer per flat (J.R. Peters). All plants were grown at 22°C in a 16:8 light:dark daily schedule, with light fluence of 62 to 74 μmol/m2/s using GroLux T5 fluorescent lights (Sylvania).
RNA Extraction, RT-PCR, and Real-Time qRT-PCR
Total RNA was prepared from agar-grown plants using the UltraClean kit according to the manufacturer's instructions (Mo Bio Laboratories). Semiquantitative RT-PCR was performed on RNA from 2-week-old plants according to standard procedures using Improm-II reverse transcriptase (Promega). PAE2 expression was measured for comparison for the RT-PCR (Downes et al., 2003). Gene-specific primers are given in Supplemental Table S1 (Integrated DNA Technologies). PCR reactions were performed on a PTC-200 thermocycler (MJ Research) with primer-specific modifications to the following program: melting step of 95°C for 5 min, variable numbers of PCR cycles of 94°C for 30 s, 52°C to 62°C for 1 min, 72°C for 1 min, and a final elongation step at 72°C for 5 min. Aliquots of RT-PCR reactions were drawn at 5-cycle intervals to ensure timing of linear phase. qRT-PCR runs were performed on RNA extracted from the blades of fifth leaves of wild type at 2-d intervals or from whole plants of wild type and OE classes 5 d after germination using a MyiQ iCycler (Bio-Rad) with SYBR Green PCR Master mix (TOYOBO). The following program was used: melting step of 95°C for 3 min, then 45 cycles of 95°C for 30 s and 60°C for 30 s. UBIQUITIN10 was used to normalize expression in qRT-PCR analyses. These qRT-PCR assays were repeated twice.
Genetic Constructs and Plant Transformation
Genetic constructs were created using the pCAMBIA 1300 binary vector as the backbone. These constructs were introduced into Agrobacterium tumefaciens strain GV3101, and the resulting strains were used to transform Arabidopsis by the floral dip method (Clough and Bent, 1998).
Resin-Embedded Sectioning
For fixation, tissue was vacuum-infiltrated with ice-cold formaldehyde-acetic acid solution (50% ethanol, 5% glacial acetic acid, 10% formalin, 35% water) for 15 min and incubated at 4°C for 3 h. The tissue was dehydrated in a series of ethanol solutions: 50%, 70%, 80%, 90%, and 100%, for at least 15 min each step, then washed twice in liquid Technovit 8100 resin (Hereus Kulzer) for 15 min for each step. The tissue was then vacuum-infiltrated with fresh resin and incubated at 4°C for 4 d, then washed in resin plus Technovit hardener II and embedded in fresh resin plus hardener in 0.2-mL plastic PCR tubes. Embedded tissue was allowed to cure for at least 1 week at 4°C before being mounted and sectioned. Then 7-μm-thick longitudinal sections were cut through tissues using a Leica RM2145 microtome.
Confocal and Environmental Scanning Electron Microscopy
For confocal imaging of root tissue, fresh plate-grown roots were immersed in 10 μg/mL propidium iodide for 1 min, washed briefly, and immediately scanned using a Zeiss LSM 510 META laser scanning confocal microscope (Zeiss) equipped with a HeNe 543-nm, 5-mW laser. For environmental scanning electron microscopy (ESEM), an FEI QUANTA 200 ESEM (FEI Company) was used on unfixed material, at 0.93 torr, 3.0 spot size, and 25-mV voltage.
Ploidy, Cell Size, and Nuclear Size Measurements
In situ nuclear DNA content measurements were performed on the fourth true leaves of 3-week-old plants. Leaves were fixed in 3:1 ethanol to acetic acid and 1 mm MgCl2, then cleared for 12 to 14 h in 95% ethanol containing 1 mm MgCl2. Nuclei were then washed twice for 15 min with phosphate buffered saline (PBS) buffer containing 1 mm MgCl2, stained with 1 μm DAPI in PBS + 1 mm MgCl2 for 15 min and again washed thrice for 15 min in PBS + MgCl2. An average of seven stained trichome nuclei and nearby guard cell nuclei from each fourth leaf of 10 fzr2-1 and 10 Columbia-0 wild-type plants was visualized through an Olympus BX60 epifluorescence microscope using a cooled CCD camera (Cascade Photometrics). The same exposure time was used for all images. Images were cropped and converted to grayscale using Adobe Photoshop 7.0. The light outputs of individual trichome and guard cell nuclei were analyzed with ImageJ (http://rsb.info.nih.gov/ij/) using a protocol similar to that of Szymanski and Marks (1998). Modifications included comparing the fluorescence of trichome nuclei only to guard cell nuclei within the same Z-series. For measurement of root nuclei, whole roots were stained with DAPI, and nuclei were imaged according to the above protocol. We measured 46 wild-type, 22 OE III, and 24 OE I root nuclei. Only cortex cell nuclei were measured, because this was the easiest cell layer to identify.
Cell external surface area measurements were obtained via ESEM micrographs of fresh tissue from fifth leaves at 500× magnification. Seven plants from each genotype were used and cell numbers were 180, 60, 131, and 279 for OE I, OE III, wild-type, and fzr2-1, respectively. The regions micrographed were adaxial surfaces halfway along the proximodistal axis of the leaf, equidistant from midvein and the leaf edge along the mediolateral axis, on both sides of the midvein. Cell areas were measured in ImageJ, tracing by hand using a Graphire drawing pad (Wacom).
Flow Cytometry
The fifth leaf of 6 3-week-old plants for each genotype was bisected just above the petiole. Each leaf was chopped continuously by hand, using a fresh razor blade, for 5 min while immersed in PI chopping buffer (1 mm MgSO4, 15 mm KCl, 5 mm HEPES, 1 mg/mL dithiothreitol, and 100 μg/mL PI). The resulting liquid was passed through a 40-μm nylon mesh to remove cellular debris. After 20 to 60 min of incubation on ice, the suspensions were run through a FACSCalibur flow cytometer (BD) and 10,000 flow cytometric events were recorded. The output was gated to eliminate signal from chloroplasts and debris.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enlarged images of wild-type (A) and AP3∷FZR2 (B) petal cells.
Supplemental Table S1. Primers used for genotyping and for RT-PCR analysis of transcript levels.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resources Center for providing the SALK_083656 (fzr2-1) and SALK_101689 (fzr2-2) mutant seeds; the University of Wisconsin Botany Department Microscope Facility for use of confocal, epifluorescent, and ESEM; CAMBIA for vectors; Dr. Brian Downes for reagents; Dr. Maria von Balthazar for sectioning expertise; and Dr. David Baum for bacterial stocks and microtome use. The confocal imaging and electron microscopy were performed at the Plant Imaging Center, Department of Botany, University of Wisconsin, Madison.
This work was supported by the University of Wisconsin Predoctoral Training Program in Genetics (grant no. 5 T32 GM07133 to Z.L.-R.), by the National Science Foundation (grant nos. IOS–0642865 and IOS–0821884 to P.H.M.), by the National Natural Science Foundation of China (grant nos. 30570933 and 30628004 to Z.L.), and by the University of Wisconsin Botany Department and Genetics Department (to Z.L.-R. and C.D.D). This is paper number 3641 of the University of Wisconsin Laboratory of Genetics.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zachary Larson-Rabin (zmlarson@wisc.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, de Vusser K, de Tavernier E, de Bock K, Inzé D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis SM, Torii KU (2007) Autonomy of cell proliferation and developmental programs during Arabidopsis above ground organ morphogenesis. Dev Biol 304 367–381 [DOI] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove D (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Day SJ, Lawrence PA (2000) Measuring dimensions: the regulation of size and shape. Development 127 2977–2987 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215 407–419 [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35 729–742 [DOI] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Tsukaya H (2007) Dissection of enhanced cell expansion processes in leaves triggered by a defect in cell proliferation, with reference to roles of endoreduplication. Plant Cell Physiol 48 278–286 [DOI] [PubMed] [Google Scholar]
- Fülöp K, Tarayre S, Kelemen Z, Horváth G, Kevei Z, Nikovics K, Bakó L, Brown S, Kondorosi A, Kondorosi E (2005) Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4 1084–1092 [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Johannesson H, Engstrom P (2001) Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HD Zip gene ATHB13. Plant Mol Biol 45 247–262 [DOI] [PubMed] [Google Scholar]
- Hase Y, Fujioka S, Yoshida S, Sun G, Umeda M, Tanaka A (2005) Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J Exp Bot 56 1263–1268 [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D (1993) cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125 1711–1721 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Misra S, Jurgens G (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76 555–566 [DOI] [PubMed] [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jugens G, Hulskamp M (2003) The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol 131 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40 77–105 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697 [DOI] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns A (1998) Auxin-dependent cell expansion mediated by overexpressed Auxin-Binding Protein1. Science 282 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Hagemann W (1991) The relationship of cell and organism in vascular plants. Bioscience 41 693–703 [Google Scholar]
- Kato N, Lam E (2003) Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J Cell Sci 116 2195–2201 [DOI] [PubMed] [Google Scholar]
- Kudo N, Kimura Y (2001) Flow cytometric evidence for endopolyploidization in cabbage (Brassica oleracea L.) flowers. Sex Plant Reprod 13 279–283 [Google Scholar]
- Kudo N, Kimura Y (2002) Nuclear DNA endoreduplication during petal development in cabbage: relationship between ploidy levels and cell size. J Exp Bot 53 1017–1023 [DOI] [PubMed] [Google Scholar]
- Lee HC, Chiou DW, Chen WH, Markhart AH, Chen YH, Lin TY (2004) Dynamics of cell growth and endoreduplication during orchid flower development. Plant Sci 166 659–667 [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4 533–539 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R, John PCL (2007) Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: implications for the control of plant growth. Plant Physiol 144 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwann T (1839) Microscopical Researches into the Accordance in the Structure and Growth of Animals and Plants. G.E. Reimer, Berlin [DOI] [PubMed]
- Serralbo O, Pérez-Pérez JM, Heidstra R, Scheres B (2006) Non-cell-autonomous rescue of anaphase-promoting complex function revealed by mosaic analysis of HOBBIT, an Arabidopsis CDC27 homolog. Proc Natl Acad Sci USA 103 13250–13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF (1997) Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90 671–681 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155 349–361 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6 1–10 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks M (1998) GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10 2047–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarayre S, Vinardell JM, Cebolla A, Kondorosi A, Kondorosi E (2004) Two classes of the Cdh1-type activators of the anaphase-promoting complex in plants: novel functional domains and distinct regulation. Plant Cell 16 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J, Hülskamp M, Gendreau E, Hofte H (1998) Endoreduplication and development: rule without dividing? Curr Opin Plant Biol 1 498–503 [DOI] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2003) Organ shape and size: a lesson from studies of leaf morphogenesis. Curr Opin Plant Biol 6 57–62 [DOI] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC (2000) SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127 3931–3940 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24 613–623 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Mészáros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bögre L, Genschik P (2004) Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Cell 16 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJH, Nowack MK, Jakoby MJ, Hülskamp M, Schnittger A (2005) Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Murakami H, Okayama H (1997) A WD-repeat protein controls the cell cycle and differentiation by negatively regulating Cdc2/B-type cyclin complex. Mol Biol Cell 8 2475–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.