Abstract
Transcriptome and metabolite profiling of rice (Oryza sativa) embryo tissue during a detailed time course formed a foundation for examining transcriptional and posttranscriptional processes during germination. One hour after imbibition (HAI), independent of changes in transcript levels, rapid changes in metabolism occurred, including increases in hexose phosphates, tricarboxylic acid cycle intermediates, and γ-aminobutyric acid. Later changes in the metabolome, including those involved in carbohydrate, amino acid, and cell wall metabolism, appeared to be driven by increases in transcript levels, given that the large group (over 6,000 transcripts) observed to increase from 12 HAI were enriched in metabolic functional categories. Analysis of transcripts encoding proteins located in the organelles of primary metabolism revealed that for the mitochondrial gene set, a greater proportion of transcripts peaked early, at 1 or 3 HAI, compared with the plastid set, and notably, many of these transcripts encoded proteins involved in transport functions. One group of over 2,000 transcripts displayed a unique expression pattern beginning with low levels in dry seeds, followed by a peak in expression levels at 1 or 3 HAI, before markedly declining at later time points. This group was enriched in transcription factors and signal transduction components. A subset of these transiently expressed transcription factors were further interrogated across publicly available rice array data, indicating that some were only expressed during the germination process. Analysis of the 1-kb upstream regions of transcripts displaying similar changes in abundance identified a variety of common sequence motifs, potential binding sites for transcription factors. Additionally, newly synthesized transcripts peaking at 3 HAI displayed a significant enrichment of sequence elements in the 3′ untranslated region that have been previously associated with RNA instability. Overall, these analyses reveal that during rice germination, an immediate change in some metabolite levels is followed by a two-step, large-scale rearrangement of the transcriptome that is mediated by RNA synthesis and degradation and is accompanied by later changes in metabolite levels.
Germination is a series of events that begins with imbibition, the uptake of water by the dry seed, followed by reinitiation of metabolic processes, elongation of the embryonic axis, and, by strict definition, terminates when part of the embryo emerges from the structures that surround it (Bewley, 1997). Germination can be divided into three phases; phases I and II are characterized by the rapid uptake of water and a plateau phase of water uptake, respectively. These phases represent a period of large metabolic change that primes the embryo to commence growth during phase III, when further uptake of water occurs (Bewley, 1997). Once the process of germination has commenced, utilization of stored reserves for energy production is necessary before the plant becomes autotrophic by establishing photosynthesis. The importance of energy metabolism in the early stages of seed germination can be seen in studies that inhibit germination, in phase II, by the use of various bioactive compounds or mutants. The alterations of transcript signatures or profiles in these studies reveal that many are associated with energy production and associated biosynthetic pathways (Carrera et al., 2007; Bassel et al., 2008).
Historically, regulation of germination has been described by the antagonistic interaction of the phytohormones abscisic acid (ABA) and GA, whereby ABA represses germination and GA promotes germination (Bewley, 1997; Holdsworth et al., 2008a). However, evidence is growing for a role of auxin during this process as well as the interaction of other phytohormones such as ethylene and brassinosteroids (Holdsworth et al., 2008a). Inhibition of transcription and translation has differential effects on germination potential. It was shown over 40 years ago that transcription was not required for de novo protein synthesis in imbibed seeds, which suggested that endogenous mRNA was utilized in early stages of the germination process (Dure and Waters, 1965). Recent studies on seed germination have shown that as many as 12,000 mRNA molecules are present in mature seeds in Arabidopsis (Arabidopsis thaliana) and barley (Hordeum vulgare; Nakabayashi et al., 2005; Sreenivasulu et al., 2008), consistent with the role of preexisting mRNA molecules playing a central role in germination. While transcriptional inhibition slows the progression of germination, radicle protrusion still occurs, although subsequent seedling growth is prevented. In contrast, inhibition of translation completely inhibits germination (Rajjou et al., 2004).
Although seed development and germination have been studied for several decades, recent advances in our understanding of these complex processes have largely resulted from the expansion of available sequence data and the establishment of large-scale -omics technologies. In particular, for the dicot model, Arabidopsis, a number of studies utilizing transcriptomic, proteomic, and metabolomic methods to investigate seed maturation, dormancy, and maturation have been published (Nakabayashi et al., 2005; Holdsworth et al., 2008a, 2008b), including one study that reports a correlation between transcript and metabolite data during the germination process (Fait et al., 2006).
In comparison, there is a relative paucity of similar studies in monocots, particularly at the whole genome level, with respect to transcriptomic and metabolomic studies. While some transcriptome studies in wheat (Triticum aestivum) and barley have been performed (Watson and Henry, 2005; Wilson et al., 2005; Sreenivasulu et al., 2008), the lack of complete genome sequence data prevents comprehensive whole transcriptome analysis, including promoter analysis once coexpressed gene sets have been identified. For example, the most comprehensive transcriptome study in monocots to date, using barley (Sreenivasulu et al., 2008), reported that cis element searches were performed in homologous rice (Oryza sativa) promoters, as this sequence information is not yet available for barley. Also, time points sampled were 24 h after imbibition (HAI) or more apart (Sreenivasulu et al., 2008), meaning that early and potentially regulatory changes in the transcriptome have not yet been thoroughly investigated in monocots.
Rice is an important food crop and is the first crop to have its genome sequenced, making it the model of choice for grass species. Several conditions established rice as the optimal choice for global germination analysis in monocots: (1) the availability of whole genome sequence information; (2) an established growth system for studying germination (Howell et al., 2006, 2007); (3) widespread functional annotation information; and (4) the availability of Affymetrix whole genome rice microarrays representing 51,279 transcripts. All of these factors together enabled comprehensive transcriptome analysis in rice over a germination time course and the direct link of coexpressed genes with upstream sequence information for identification of potential regulatory sequence elements. Sampling transcripts in dry seeds (0) and 1, 3, 12, and 24 HAI and of metabolites at 0, 1, 3, 6, 12, 24, and 48 HAI has allowed a detailed examination of germination in rice. Furthermore, this study enables the investigation of the roles of transcriptional and posttranscriptional processes and whether changes in transcript levels drive changes in metabolites during this essential phase of plant growth and establishment.
RESULTS
Transcriptome and Metabolite Profiling of Early Stages of Rice Germination
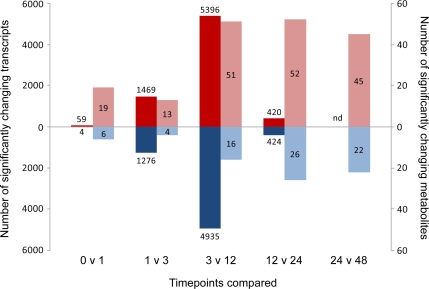
We have previously characterized changes in water content and metabolic activity in rice embryos during germination up to 48 HAI and have observed the expected triphasic mode of water uptake with concomitant increases in oxygen uptake (Howell et al., 2006). Using this same experimental system, global changes in transcript levels were determined 0, 1, 3, 12, and 24 HAI using the Affymetrix Rice GeneChip, consisting of 57,381 probe sets representing 51,279 transcripts. Microarrays were performed in triplicate for each time point, and after normalization, analysis of the data revealed that the correlation between the replicates for each time point was greater than 0.98. The total number of probe sets for analysis was reduced by removing ambiguous probe sets and those that were not called “present” in at least two replicates at one time point, resulting in a final present set of 24,150 transcripts (Supplemental Table S1). Of these, over 17,000 transcripts were present prior to imbibition (i.e. representing the mRNA stored in the dry seed). Differential expression analysis (with false discovery rate correction and a stringent cutoff of P < 0.01) revealed that 76% (18,372) of these transcripts changed in abundance over the time course and 67% (16,487) changed relative to 0 HAI. (Supplemental Fig. S1; Supplemental Table S1). When successive time points are compared, there were relatively few changes in the first hour (59 up, four down), with most changes observed between 3 and 12 HAI (5,396 up, 4,935 down) and between 1 and 3 HAI (1,469 up, 1,276 down), while the number of changes between 12 and 24 HAI was considerably lower (420 up, 424 down; Fig. 1). It was apparent that almost as many transcripts decreased in abundance as increased at almost all time point comparisons.
Figure 1.
Summary of the number of significant changes in transcripts and metabolites between successive time points during rice germination. Transcript and metabolite profiling were performed on rice embryo tissue samples collected at various time points during germination (0, 1, 3, 12, 24, and 48 HAI). Changes in the abundance of 24,150 transcripts and 126 metabolites were determined, and statistical analysis was performed to evaluate significant differences between all possible combinations of time points (Supplemental Fig. S1). Comparison of successive time points for significantly up-regulated (red) and down-regulated (blue) transcripts (dark red and blue; left axis) and metabolites (light red and blue; right axis) revealed differences in the timing of significant alterations in the transcriptome and metabolome. The numbers of significantly changing transcripts and metabolites for each comparison are given above and within the columns, respectively. nd, Not determined.
Metabolite analysis was performed on the same samples used for microarray analysis and additional samples collected 6 and 48 HAI. A total of 126 unique metabolites were detected in the rice embryo samples, and of these, 66 could be identified based on matching to previously run standards (Supplemental Table S2A). Statistical analysis of metabolite abundance revealed that most (93%) of the 126 metabolites detected showed significant (P < 0.05) changes in abundance between at least two time points sampled during the time course, and of the 66 metabolites identified, all were found to show significant changes in abundance (Fig. 1; Supplemental Table S2B). Although a number of significant changes in metabolite abundance were observed just 1 HAI (25 of the 126 metabolites displayed significant changes between 0 and 1 HAI), the differences between 1, 3, and 6 HAI were more subtle compared with the large changes in the transcriptome observed from 1 to 3 HAI (Fig. 1; Supplemental Table S2B). In contrast, large overall changes in metabolite profiles were observed from 12 HAI onward (Fig. 1), with more than 50 of the metabolites displaying a significant change in abundance at 12 HAI or later (Fig. 1; Supplemental Table S2A). Overall, examination of the changes in metabolites and transcripts revealed a rapid change in metabolite levels within 1 HAI, which preceded the large changes in transcript abundance at 3 and 12 HAI and was followed by further changes in metabolites at 12 HAI or later.
Comparing Patterns of Specific Metabolites and Transcripts during Germination
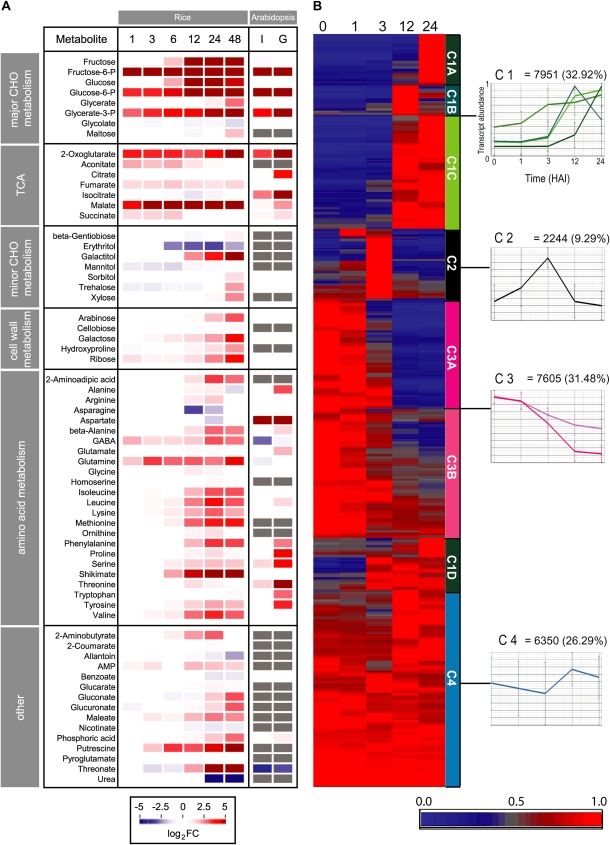
The striking changes in metabolite levels that occurred just 1 HAI were predominantly associated with major carbohydrate metabolism (Fig. 2A; Supplemental Table S2B). Fru-6-P, Glc-6-P, and glycerate-3-phosphate increased 7- to 42-fold between 0 and 1 HAI and were also observed to increase at all time points thereafter. Other metabolites found to rapidly increase included the tricarboxylic acid (TCA) cycle intermediates 2-oxoglutarate, aconitate, fumarate, malate, and succinate, with increases ranging from 2.7- to over 16-fold. This suggests that there is an immediate increase in the activity of glycolysis and the TCA cycle that facilitates early, energy-demanding processes. While most of the changes in amino acids were seen to occur later in the time course, γ-aminobutyric acid (GABA) and Gln were notable exceptions, displaying 4.2- and 3.5-fold increases at 1 HAI (Fig. 2A; Supplemental Table S2B). In addition to increases in several amino acids (Ile, Leu, Lys, Met, Phe, Ser, Tyr, and Val), changes in the metabolite profiles at later stages of germination also included increases in sugars (Fru, Glc, and maltose), compounds associated with cell wall metabolism (Ara, Gal, Hyp, and Rib), and minor carbohydrate metabolism (galactitol, sorbitol, trehalose, and Xyl).
Figure 2.
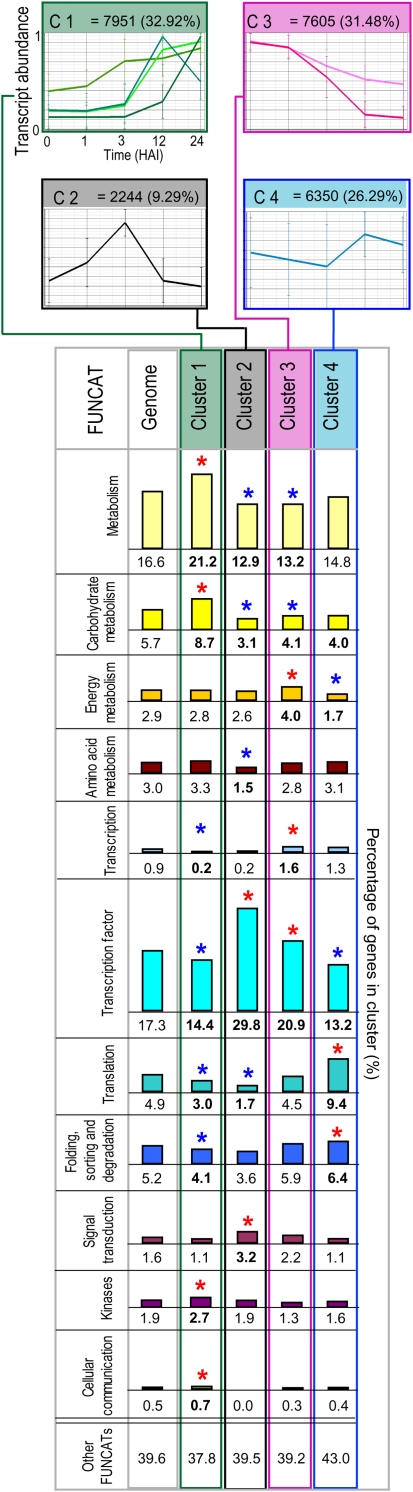
Profiles of known metabolites and hierarchical clustering of differentially expressed genes during rice germination. A, Changes in the levels of all identified metabolites were calculated as fold changes relative to the 0-HAI time point and log transformed. Changes are represented as a false color heat map where the color saturates at a log2 false color (FC) value of 5 (i.e. a 32-fold change). Data from a study performed using whole Arabidopsis seeds (Fait et al., 2006) are included for comparison, where I represents seeds imbibed for 72 h at 4°C in the dark relative to dry seeds and G indicates a comparison of seeds imbibed for 72 h at 4°C in the dark followed by 24 h of growth under germinative conditions (21°C in the light) relative to dry seeds. White coloring indicates no significant change, and gray coloring indicates that a metabolite was not measured. The fold changes for all metabolites detected and associated P values are shown in Supplemental Table S2B. B, From microarray analysis, all probe sets that were called present at a minimum of one time point were normalized to the highest level of expression over the time course of the study and hierarchically clustered using average linkage based on Euclidian distance. Four primary clusters were defined: cluster 1 (green), transcripts that increased in abundance over the time period examined; cluster 2 (black), transcripts that were low or absent at 0 HAI, peaked at 1 or 3 HAI, and then declined in abundance; cluster 3 (pink), transcripts that declined in abundance over the time period examined; cluster 4 (blue), transcripts that displayed relatively stable levels of abundance throughout the time course. Subclusters of clusters 1 and 3 are defined by differences in the time points at which changes in transcript levels occurred. For all clusters, a graph showing the average expression level is presented. Fold changes and their associated P values for all probe sets can be found in Supplemental Table S1.
To compare these metabolite patterns with profiles observed for the 24,150 transcripts detected during rice germination, transcript abundance data were normalized to the highest value for each transcript and then hierarchically clustered, resulting in four main types of transcript profile patterns (Fig. 2B). Cluster 1 represents just under one-third of all expressed genes and is characterized by transcripts that have relatively low and stable levels at early stages of germination and then increase over the time course examined. Cluster 1 was subdivided into four subgroups (A–D) based on when the increase in transcript abundance was observed: cluster 1A increases from 12 to 24 HAI; cluster 1B increases from 3 to 12 HAI, followed by decreases from 12 to 24 HAI; cluster 1C increases between 3 and 12 HAI and remains high at 24 HAI; and cluster 1D increases after 1 or 3 HAI (Fig. 2B). Cluster 2 (black) was unique in that the transcript abundance profiles peaked in abundance after 1 or 3 HAI and then decreased to low levels again from 12 HAI (Fig. 2B), suggesting that a distinct regulatory process has occurred that transiently affects the transcript abundance of over 2,000 genes. Furthermore, the transient but dramatic increases in the transcripts that constitute cluster 2 precede the majority of the increases in transcripts observed for cluster 1, which occur at 12 HAI or later (Fig. 2B). Cluster 3 (pink) is defined by transcripts that decrease throughout the time course of the study, representing almost one-third of all expressed genes, and can be divided into two subgroups: 3A, in which the profiles showed a general decrease in transcript abundance, starting after 1 HAI and continuing to 12 HAI; and 3B, in which the decrease was not as dramatic as that observed for cluster 3A. Cluster 4 (blue) comprises just over one-quarter of all expressed genes and showed relatively constant transcript levels across the time course (Fig. 2B). Interestingly, cluster 1C and cluster 3A are practically mirror images, in that they both include around 3,500 genes, and while cluster 1C shows an increase at 3 HAI, cluster 3A displays a corresponding decrease.
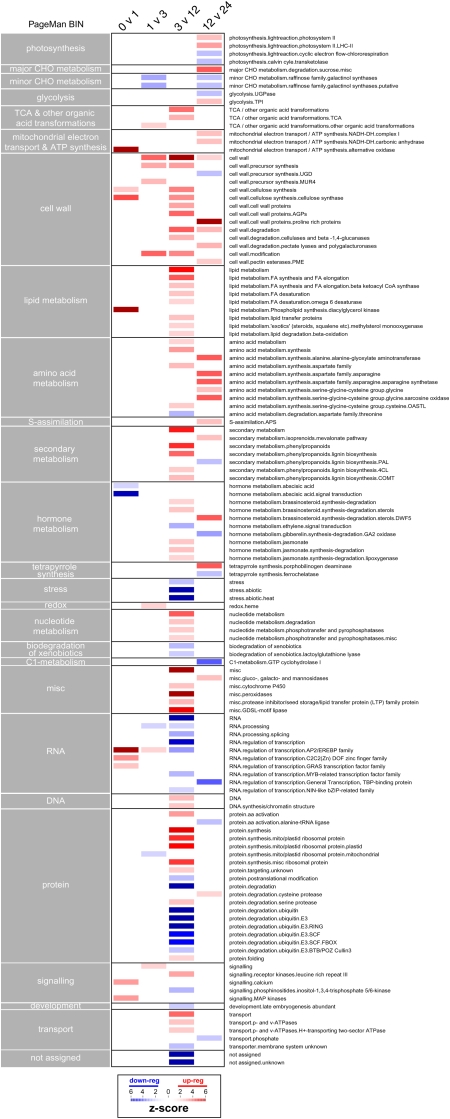
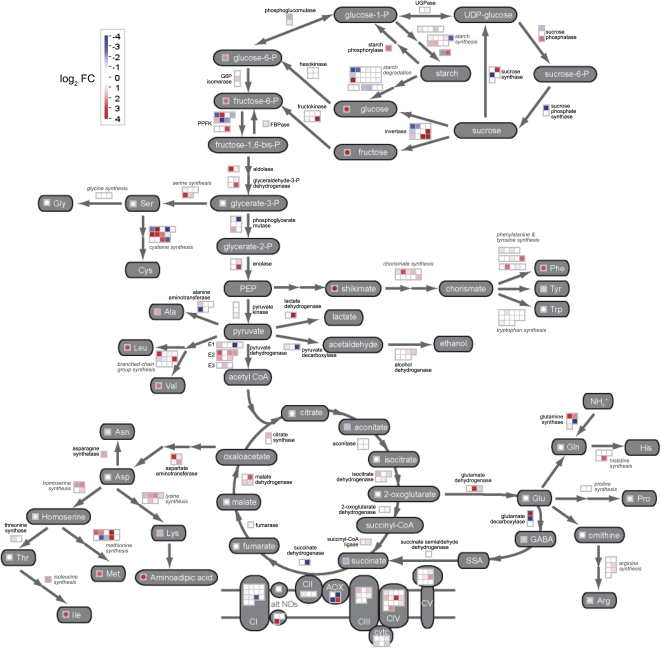
To understand the significance of these distinct patterns of transcript abundance and their relationship to the metabolome changes, three types of analysis were conducted that each provided a different insight into a molecular understanding of the germination process in rice. The first analysis was performed using the PageMan (Usadel et al., 2006) and MapMan (Thimm et al., 2004; Usadel et al., 2005) tools adapted for use with rice microarray data (see “Materials and Methods”). This type of analysis was performed by comparing only significant changes between successive time points and reveals which functional categories are significantly up- or down-regulated. PageMan analysis revealed that a variety of cellular processes were affected over the germination process and confirmed that the greatest number of significant changes were observed between 3 and 12 HAI (Fig. 3). Early changes (0 versus 1 HAI) included signaling processes involving transcription regulation, mitogen-activated protein kinases, and calcium. Further analysis using MapMan and selected time points (0 versus 3 HAI and 3 versus 12 HAI; Supplemental Fig. S2) also provided further insight into the signal transduction pathways that where utilized. For example, it showed that transcripts of receptor kinases were up-regulated at early stages of germination (0–3 HAI), while later changes (3–12 HAI) involved an up-regulation of transcripts associated with the brassinosteroid and jasmonate pathways. Auxin-responsive transcription factors were also up-regulated at these later time points, while, in general, components involved in protein degradation and modification were repressed (Supplemental Fig. S2). However, early changes in transcript levels were not restricted to regulatory processes, as transcripts encoding proteins involved in cellulose and phospholipid synthesis, as well as one isoform of the alternative oxidase (AOX1a), were found to be up-regulated, while transcripts associated with abscisic acid signal transduction were down-regulated (Fig. 3). Large changes in the transcriptome from 3 to 12 HAI were associated with a general up-regulation of transcripts encoding components involved in the following cellular processes: cell wall metabolism, lipid metabolism, nucleotide degradation, amino acid synthesis, carbohydrate metabolism (TCA cycle), jasmonate synthesis, cellular transport, organellar protein synthesis, and aspects of secondary metabolism such as isoprenoid and phenylpropanoid biosynthesis (Fig. 3). These observations are further supported by a more specific comparison of metabolism using MapMan. This analysis showed up-regulation of several biosynthetic pathways, such as cellulose synthesis, cell wall synthesis, cell wall modification, tetrapyrrole synthesis, fatty acid synthesis, and β-oxidation (Supplemental Fig. S3). Although there were fewer changes associated with later time points (12 versus 24 HAI), these were associated with central processes such as photosynthesis, Suc degradation, mitochondrial electron transport, tetrapyrrole synthesis, brassinosteroid metabolism, and amino acid synthesis (Fig. 3), indicating that central metabolic processes are being maintained and/or induced in preparation for seedling establishment during late stages of germination. Furthermore, for some of these pathways, metabolite components were also identified, and increases in these components, such as those involved in amino acid and cell wall metabolism (Fig. 2A), correlate with general changes in transcript levels (Fig. 3). To further investigate major carbon and amino acid metabolism, a custom MapMan pathway image was generated. Fold changes in transcript (3 versus 12 HAI) and, where possible, metabolite levels (6 versus 24 HAI) were plotted simultaneously (Fig. 4). It was found that by displaying the data in this manner and introducing a time lag between the transcript and metabolite changes, there was a better correlation between changes, particularly with regard to the induction of transcripts involved in amino acid synthesis and the levels of the amino acids themselves (Phe, Tyr, Ala, Leu, Val, Lys, Met, Ile, and Arg). Thus, although metabolomic analysis is not yet as comprehensive as transcriptomic analysis, it suggests that the extensive changes in the transcriptome between 3 and 12 HAI drive the later changes observed in metabolite profiles.
Figure 3.
PageMan analysis of the microarray data over the rice germination time course. Significant fold changes in transcript levels between adjacent time points were log transformed and analyzed using the PageMan tool. Wilcoxon statistical analysis with Benjamini-Hochberg false discovery rate control was performed to determine significantly different gene categories. Nonsignificant categories were collapsed for display. Statistical differences are represented by a false color heat map (red = up-regulated; blue = down-regulated) where a z-score of 1.96 represents a false discovery rate-corrected P value of 0.05.
Figure 4.
Parallel display of transcripts and metabolites for starch-Suc metabolism, glycolysis, the TCA cycle, GABA shunt, mitochondrial respiratory chain, and amino acid metabolism. Significant fold changes in transcripts and metabolites were log transformed and displayed on a custom pathway picture using the MapMan tool. Where possible, metabolite changes are indicated in the circles next to the corresponding metabolite name (gray boxes) and correspond to a comparison of 6 and 24 HAI. Enzymatic conversions between metabolites are indicated by arrows and enzyme names. Changes in transcripts encoding these enzymes are indicated in the boxes next to the enzyme names and correspond to the 3- versus 12-HAI comparison. In most cases, the enzymes involved are encoded by a small gene family. However, in some cases, individual enzymes are not distinguished and a more general classification of the contributing transcripts is indicated in italics (e.g. starch degradation). This is also the case for components of the mitochondrial electron transport chain (CI–CV), where transcript levels for different nucleus-encoded subunits are presented. For both metabolites and transcripts, changes are represented by shading, where the color saturates at a log2 false color (FC) value of 4 (i.e. a 16-fold change).
The above analyses reveal the characteristics of statistically significant changes between successive time points. Thus, it primarily gives insights into the changes that are occurring in clusters 1 and 3, where large fold changes of many transcripts are occurring. It is not informative for sets of genes that do not change (i.e. cluster 4) and also may miss some changes that occur in clusters with smaller numbers of genes (i.e. cluster 2). Thus, a second analysis approach was carried out on changes in transcripts based on all transcript profiles (i.e. the 24,150-gene set; Fig. 2B) and the functional categories of the encoded proteins. Differences were determined by calculating z-scores to test if the percentage of a particular category was significantly higher or lower (P < 0.01) than in the whole genome (Fig. 5; Supplemental Fig. S4; Supplemental Table S3). Cluster 2, characterized by transient increases in abundance at early stages of germination (1 and 3 HAI), was found to contain a significantly higher proportion of transcripts encoding transcription factors and proteins involved in signal transduction and was underrepresented in several categories of metabolism (Fig. 5). Cluster 4, which displayed relatively constant profiles over the 24-h time period, was found to have a higher proportion of transcripts associated with translation as well as protein folding, sorting, and degradation, which suggests a consistent requirement of the proteins involved in these functions (Fig. 5; Supplemental Fig. S4; Supplemental Table S3). Furthermore, for transcripts comprising clusters 1 and 3, the findings of the PageMan/MapMan analysis were supported by this type of approach.
Figure 5.
Functional categorization of the transcripts grouped into each cluster. Of the 24,150 transcripts that were present at any one time point, a functional classification could be ascribed to over 10,000, as outlined in “Materials and Methods.” The breakdown of the genome as well as the four clusters defined in Figure 2B are shown. The frequency of transcripts in each FUNCAT was calculated as a percentage of the cluster and compared with the percentage of the genome in that FUNCAT. Functional groups that were found to be overrepresented (red asterisks) or underrepresented (blue asterisks) are listed for each cluster, as determined by the z-score test with a confidence of P < 0.01. Only the FUNCATs that changed significantly are shown (for a complete list of all FUNCATs in all clusters, see Supplemental Fig. S4).
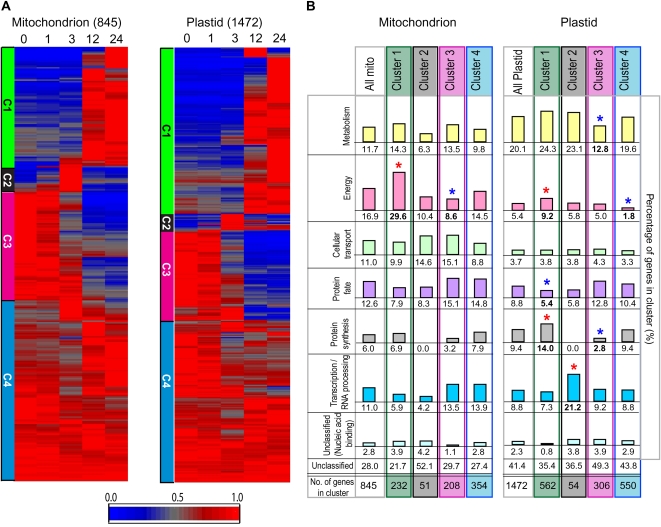
Third, sequential changes in metabolic organelle function (plastids, mitochondria, and peroxisomes) were investigated during germination. In order to determine if transcripts that encode organelle proteins changed in a coordinated manner compared with that observed for all transcripts (Fig. 2B), subsets of transcripts for genes encoding organelle proteins were reanalyzed by clustering analysis. Four clusters could be clearly defined for transcripts that encoded proteins located in mitochondria, plastids, and peroxisomes based on similar temporal changes in transcript abundances (Fig. 6A; Supplemental Fig. 5) compared with what was observed when the abundances of all transcripts of the array defined as present were clustered using identical parameters. Functional categorization analysis of the organelle cluster sets was used to determine which categories were overrepresented or underrepresented in each cluster (Fig. 6B). For both mitochondrial and plastid sets, transcripts encoding proteins involved in energy were found to be overrepresented in cluster 1 and underrepresented in cluster 3 (mitochondrial) and cluster 4 (plastid). This enrichment of energy functions in cluster 1 correlates with the requirement for large amounts of energy in the early stages of germination. For the plastid set, transcripts associated with protein synthesis were also overrepresented in cluster 1, while transcripts associated with protein fate were underrepresented (Fig. 6B). This may correspond with the order of processes that occur in organelles that have their own genome. Previous studies investigating mitochondrial biogenesis during rice germination revealed that the transcripts encoding for import components (protein fate) appear first, followed by the other organelle-localized proteins, which can only enter the organelle via these import components (Howell et al., 2006). Therefore, transcripts encoding protein fate components are required early and hence would not steadily increase, as in cluster 1, whereas the transcripts encoding protein synthesis components increase over time, facilitating specific protein production within the plastid. With regard to cluster 2, transcripts involved in transcription and RNA processing were overrepresented in the chloroplast set, consistent with the whole genome analysis (Fig. 2B). The essential role of mitochondria and plastids during seed development, early germination, and seedling growth is reflected in the observation that a large proportion of the transcripts are not significantly changing in abundance over the first 24 HAI (cluster 4). These observations are consistent with the fact that upon imbibition, immediate changes in metabolites occur, due to the presence of significant metabolic capacity of both organelles encoded by transcripts in cluster 4. Later changes in metabolites only occur 12 HAI or later, parallel to cluster 1, enriched in genes encoding energy functions in both mitochondria and plastids.
Figure 6.
Analysis of the transcript abundance of genes encoding proteins located in mitochondria and chloroplasts. A, Hierarchical clustering of 845 genes defined to encode mitochondrial proteins and 1,472 genes defined to encode plastid proteins, divided into four clusters as outlined in Figure 2. B, Functional categorization of the proteins encoded by the genes in each cluster. The breakdown of the functional categorization in each cluster (C1–C4) and the percentage of genes are shown. Asterisks indicate significant differences (based on z-scores with P < 0.01) compared with the total organelle sets. The number of genes, percentage breakdown, and significance scores in each cluster are shown in Supplemental Table S5.
We have previously suggested a sequential assembly of mitochondria during germination based on the examination of a limited number of genes (Howell et al., 2006), and that sequential pattern is supported by the analysis of the larger set of genes in this study (Supplemental Table S5B). For example, transcript abundance of genes involved in protein import and organelle gene transcription (e.g. the mitochondrial RNA polymerase) is relatively high in dry seeds and at early germination stages and either remains high or declines (i.e. cluster 3 or 4). In contrast, many of the transcripts encoding components associated with organellar protein synthesis increase at 3 HAI, while transcripts encoding components of the TCA cycle and the respiratory chain increase at 12 HAI (i.e. clusters 1B and 1A, respectively). Notably, cluster 2 of the mitochondrial set includes transcripts encoding membrane transport proteins, including phosphate and oxoglutarate/malate carriers, Graves disease protein, a transporter necessary for the accumulation of mitochondrial coenzyme A (Prohl et al., 2001), and proteins annotated as uncoupling proteins. In Arabidopsis, these proteins have been functionally shown to transport a variety of metabolites, including the components of the malate/oxalocetate shuttle, which is an important link between mitochondrial and cytosolic metabolism (Palmieri et al., 2008). Finally, it was also seen that for some proteins that are encoded by small gene families and are involved in mitochondrial metabolism (e.g. the E1α subunit of the pyruvate dehydrogenase complex and cytochrome c), one isoform increased over the time period examined while another decreased (Supplemental Table S5B), suggesting that there may be a switch in the isoform utilized during seed maturation versus germination processes.
In combination, these three analysis approaches revealed an almost immediate change in the metabolome, followed by a two-step large-scale rearrangement of the transcriptome featuring metabolic organelle biogenesis and followed by increases in amino acids and components involved in cell wall and carbohydrate metabolism. However, this analysis does not explain what the switch or driver was for these phases in the germination process.
Transient Changes in the Transcriptome Indicate That 3 h May Represent a Specific Switch Point in the Germination Process
The above analysis of overrepresented and underrepresented functional categories revealed that transcription factors are underrepresented in clusters 1 and 4 (i.e. transcript levels that increase or remain stable) but are overrepresented in clusters 2 and 3 (i.e. transcript levels that increase only transiently or decrease; Fig. 5; Supplemental Table S3). Given that the transcription factors in cluster 3 are characterized as having their highest levels in the dry seeds before decreasing over the germination period, these putatively represent regulators involved in processes associated with seed maturation and desiccation. These transcripts appear to be stored in the dry seeds and then decay as the germination process proceeds. However, the transcription factors contained in cluster 2 are at low levels in the dry seeds and are only transiently expressed at 1 or 3 HAI and, thus, may represent an important regulatory switch that may then drive the changes in transcript abundance that occur later, particularly with respect to increases in transcript abundance represented in cluster 1.
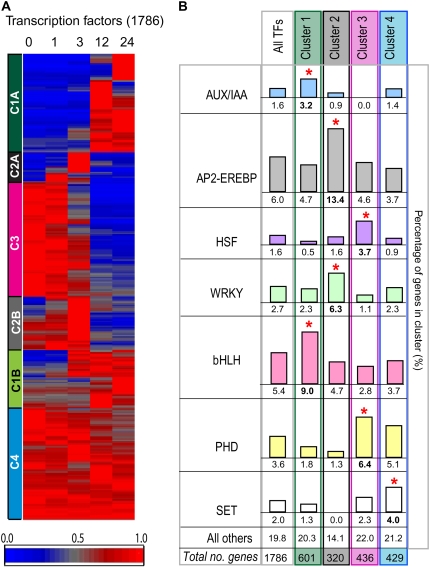
Given these interesting observations, we performed further analysis on the rice transcription factor set. A comprehensive list of rice transcription factors was collated from various databases and studies (as described in “Materials and Methods”), and it was found that transcripts for 1,786 of these were detected in at least one time point of this study. Their transcript profiles were analyzed by hierarchical clustering (Fig. 7A), and for each cluster type, the proportions of the different transcription factor families were analyzed (Fig. 7B; Supplemental Table S4B). Interestingly, it was found that there was a bias in the types of transcription factors occurring in each cluster. Cluster 1 had a higher proportion of AUX/IAA and basic helix-loop-helix families, while cluster 4 was enriched in the SET family transcription factors (Fig. 7B). These findings are consistent with the observation that members of the AUX/IAA family have previously been associated with GA and auxin signaling pathways during germination in barley (Sreenivasulu et al., 2008). Previous studies have revealed a role for SET family transcription factors in histone methylation (Malagnac et al., 2002; Xiao et al., 2003); thus, consistent expression of these family members suggests a constitutive epigenetic role of the SET family members in plant development.
Figure 7.
Analysis of changes in transcript abundance for genes encoding transcription factors. A, Hierarchical clustering of 1,786 transcription factors that were present at a minimum of one time point, divided into four clusters as in Figure 3. B, Analysis of the transcription factors by family present in each cluster. Overrepresentation is indicated by red asterisks. The frequency of transcripts, percentage breakdown in each cluster, and the significance score are shown in Supplemental Table S4B.
In contrast, cluster 2, characterized by a transient peak in expression at 3 HAI before decreasing, was found to be enriched in AP2-EREBP and WRKY family members (Fig. 7B). AP2 family members are known to play an important role in ABA signaling and in water uptake/drought response, with mutants of an AP2-EREBP family member in Arabidopsis showing increased water loss (Song et al., 2005). This transient expression may highlight an important role of AP2 transcription factors in water uptake and ABA signaling during the phases of germination. A role for WRKY family members in GA signaling has also been proposed following the expression patterns observed for WRKY family members in barley (Sreenivasulu et al., 2008), and an overrepresentation of WRKY transcription factors in cluster 2 relative to other transcription factors suggests that they may also play a role in germination processes in rice. Lastly, HSF and PHD family members were overrepresented in cluster 3, which showed high expression in dry seeds and decreasing expression over time (Fig. 7B). HSF family members have long been associated with protein folding and stress response; thus, their role in early germination appears critical, as large numbers of proteins begin production over the germination time course (Guo et al., 2008). The identification of specific genes encoding transcription factors displaying distinct temporal expression patterns provides a way to identify putative regulators that meditate the transition from dormancy to early seedling growth after imbibition.
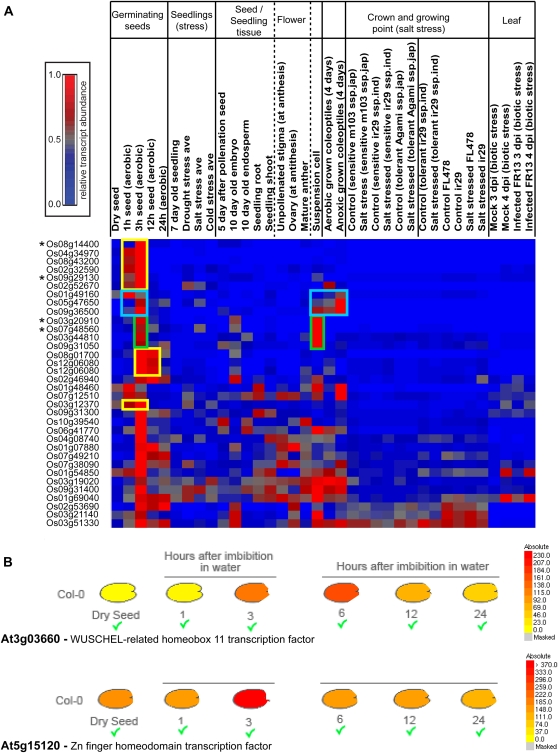
Transcription factors identified as belonging to cluster 2 (Fig. 7A) may mediate changes in transcript levels observed later in the time course (i.e. increases in the transcript abundance observed for over 6,000 genes in cluster 1). To determine if these were specific to the process of germination, we analyzed the expression of transcription factors across publicly available rice Affymetrix microarray data. These included analyses of over 30 microarrays from different tissues and stress treatments, and following normalization, all data were made relative to maximum expression across all arrays (detailed in “Materials and Methods”). The 117 transcription factors comprising cluster 2A were then examined closely across the compiled normalized data from this study and the public data, and 34 transcription factors were identified that reached at least 70% of maximum expression levels at 1 or 3 HAI when all available rice array data were analyzed (Fig. 8). Interestingly, nine of these were found to be exclusively expressed at these early time points during germination and were absent or at very low levels across all the other arrays analyzed (Fig. 8A, yellow boxes). It is important to point out that three of these belonged to the AP2-EREBP family, which further supports our conclusion on the importance of this family in regulating water uptake and ABA signaling specifically during germination. Furthermore, it was interesting that of the 1,786 transcription factors, only two belonged to the AB13/VP1-2 family, and both of these fell into the group of nine genes expressed, almost uniquely, during germination. ABI3/VP1 family members are known to play a role as intermediaries in regulating ABA-responsive genes (Lazarova et al., 2002); therefore, this distinct expression pattern suggests that these two transcription factors are likely to play a critical role during early germination in rice. Other putative “germination-specific” transcription factors showed some limited expression across other tissues/treatments, with three, including one AP2-EREBP family member, only expressed in coleoptiles, particularly under anoxic conditions (Fig. 8A, blue boxes), and four only expressed in suspension cells (Fig. 8A, green boxes).
Figure 8.
Analysis of genes encoding transcription factors that displayed germination-specific expression. A, Analysis of the expression profiles of 34 transcription factors that displayed between 70% and 100% of their maximum expression at 1 and 3 HAI in the germination time course, compared with publicly available array data for a variety of rice tissues and treatments. Boxed in yellow are transcription factors that appeared only to be induced during germination (i.e. in this study). Boxed in blue are transcription factors only expressed during germination, coleoptiles, or suspension cells, and boxed in green are transcription factors only expressed in suspension cells and in this study. Asterisks indicate genes for which the Arabidopsis homologs have germination-specific expression (see B). B, inParanoid (Remm et al., 2001) and GreenPhylDB (Conte et al., 2008) were used to identify Arabidopsis homologs for the rice transcription factors defined as germination specific (see A). Their expression profiles in seed germination were investigated using the Arabidopsis eFP browser (Winter et al., 2007). Two Arabidopsis transcription factors that showed transient and germination-specific expression were identified, including a WUSCHEL-related homeobox transcription factor (At3g03660) homologous to rice homeobox transcription factors encoded at the loci Os08g14400, Os03g20910, and Os07g48560 and a zinc finger homeodomain transcription factor (At5g15120) homologous to the rice zinc finger homeodomain transcription factor Os09g29130.
By searching for Arabidopsis homologs of the “germination-specific” transcription factors identified in this study and verifying their expression profiles using the eFP browser (Winter et al., 2007), we successfully identified two putative germination-specific transcription factors in Arabidopsis (Fig. 8B). The WUSCHEL-related homeobox 11 (At3g03660) transcription factor displays a transient increase in expression at 3 and 6 HAI in Arabidopsis seeds and is homologous to three of the transiently expressed rice homeobox transcription factors (Os08g14400.1, Os03g20910.2, and Os07g48560.1; Fig. 8B; Supplemental Fig. S6). ZPHD3/ATHB30, a zinc finger homeodomain transcription factor (At5g15210), also displays a transient increase at 3 HAI in Arabidopsis seeds and is homologous to the transiently expressed rice zinc finger homeodomain transcription factor (Os09g29130.1; Fig. 8B; Supplemental Fig. S6). This suggests the presence of common regulators of germination processes in both dicots and monocots. However, as members of the same family of transcription factors can have diverse roles, it cannot yet be concluded that these homologous transcription factors play identical roles in germination in rice and Arabidopsis. The similar temporal expression profiles as well as sequence similarity at the whole protein sequence level (Supplemental Fig. S6) suggest some common roles and clear targets for future investigations.
Transcripts Displaying Similar Profiles during Germination Share Common Sequence Motifs
Over 17,000 transcripts were observed in dry seeds, and over the germination time course, more than 18,000 of the 24,150 transcripts present in total were found to significantly change in abundance. A number of peaks in transcript abundance were observed, at 1 and 3 HAI (cluster 2) and 12 HAI (cluster 1B), while some transcripts present in dry seeds were observed to decrease (cluster 3; Fig. 2B). These changes occurred within a 24-h period, suggesting several regulatory steps. In order to uncover the regulatory processes that caused these changes, searches for the presence of common sequence elements in the promoter regions or 3′ untranslated regions (UTRs) were carried out. As outlined in “Materials and Methods,” 10 sets of genes varying in number from five to 90 were examined for sequence elements (Supplemental Table S6A). Ten sets of these genes peaked at one time point, where a peak was defined as having a transcript abundance of 1.0 (100%) at the peak time point (0, 1, 3 12, or 24 HAI) with less than 50% transcript abundance at all other times examined. Groups examined included the mitochondrial (3 and 24 HAI), plastid (3, 12, and 24 HAI), and transcription factors (0, 1, 3 12, and 24 HAI) sets (Supplemental Table S6, B and C). The mitochondrial and plastid sets did not have any transcripts that “peaked” at 0 and 1 HAI (and 12 HAI for the mitochondrial set). Searches identified a number of conserved elements in each group (Supplemental Table S6B). The transcription factor set that peaked in expression at 3 HAI contained two elements that occurred in all 51 genes. As might be expected, there was also some overlap between the elements that occurred in the different groups that peaked at the same time, and these are reflected in the color of the elements that contain a common core sequence (Supplemental Table S6C). For example, for the transcripts peaking at 3 HAI (in the plastid and transcription factors sets), the corresponding genes were found to contain the helix-turn-helix and BBr/BPC/ARF elements.
Five distinct core sequence elements were found to occur within the different groups (above), indicated by color (two related elements in purple), with variations or reverse complements shown (Supplemental Table S6C). Elements that occurred in 70% or more of the sequences from the sets above (Supplemental Table S6, B and C) were taken and searched in the larger genome sets according to expression criteria (i.e. peak expression at one time point and less than 50% at all other time points; Table I). Sequence elements in the 1-kb promoter region were found to be significantly enriched at all time points except 0 HAI (Table I). Transcripts that peaked at 24 HAI contained six elements that were significantly underrepresented and six that were overrepresented, and of these, three were unique to this time point (Table I; Supplemental Table S6D). Transcripts that peaked at 3 HAI had seven elements overrepresented, the greatest number of elements overrepresented in any group, and one element underrepresented (Supplemental Table S6D). Interestingly, two of the elements overrepresented at 3 HAI were underrepresented at 24 HAI.
Table I.
Comparison of the presence of the putative motifs within the genome and the subsets that showed maximum expression at a given time point
The sequences analyzed were all 1-kb upstream regions (66,710) and all 3′ UTR sequences (3,027) obtained from the full genome sequence information files from TIGR. For the 1-kb upstream and 3′ UTR genome sets, the number of sequences in which the motif occurred (Freq.) and the corresponding percentage (%) of all sequences that this represents are shown. A z-score analysis was carried out (Supplemental Table S6C), and the putative motifs found to be significantly overrepresented and underrepresented (+ and − at P < 0.01) are shown next to the percentage of sequences in which the motif occurred. For the putative 1-kb upstream motifs, asterisks indicate that these motifs partially/fully match known rice elements found in the Rice Cis-Element Search and/or PlantCare databases. Previous analysis of 3′ UTR sequences (Ohme-Takagi et al., 19932; Narsai et al., 20071) have suggested the involvement of these motifs in mRNA stability. Elements that contain some overlapping bases are indicated by the same superscript letter.
| Sequences Analyzed | Putative Motif | Genome
|
0-h Peaking | 1-h Peaking | 3-h Peaking | 12-h Peaking | 24-h Peaking | |
|---|---|---|---|---|---|---|---|---|
| Freq. | % | |||||||
| 1-kb upstream regions | aAAAAAAAA* | 20,377 | 30.50 | +44.90% | +40.80% | |||
| bTTTTTTTT* | 20,369 | 30.50 | +37.50% | |||||
| cCACCAC* | 20,002 | 30.00 | +53.30% | +38.80% | +39.80% | +35.10% | ||
| cACCACC | 18,762 | 28.10 | +56.70% | +38.00% | +32.90% | |||
| dGGTGGT | 15,786 | 23.70 | −20.80% | |||||
| eGCCGCC* | 23,601 | 35.40 | +43.80% | −28.00% | −26.30% | |||
| eCGCCGC | 24,318 | 36.50 | +46.10% | −28.00% | −28.50% | |||
| fGCGGCG* | 21,936 | 32.90 | −22.30% | |||||
| fGGCGGC | 21,742 | 32.60 | −25.20% | −21.20% | ||||
| gGGAGGG* | 20,008 | 30.00 | −23.40% | −24.40% | ||||
| gGAGAGA | 23,541 | 35.30 | ||||||
| hTCTCTC* | 26,217 | 39.30 | +47.70% | +47.20% | +45.70% | |||
| hCCCTCC | 23,461 | 35.20 | +44.00% | +43.70% | ||||
| hTCCTCT | 25,331 | 38.00 | +46.60% | +46.50% | +43.30% | |||
| No. of sequences analyzed | 66,710 | 27 | 30 | 384 | 254 | 1,231 | ||
| 3′ UTRs | bTTTTTT1 | 1,058 | 35.00 | +69.20% | ||||
| TTATTG | 435 | 14.40 | +42.30% | |||||
| dGCTGGT | 257 | 8.50 | +30.80% | |||||
| ATTTAT2 | 597 | 19.70 | +38.50% | |||||
| GAATAA1 | 373 | 12.30 | +31.60% | |||||
| No. of sequences analyzed | 3,027 | 2 | 3 | 26 | 19 | 79 | ||
When analyzing changes in transcript abundance, it is important to consider the role of mRNA degradation, particularly when it is evident that dramatic decreases in transcript abundance are occurring for large groups of transcripts after they peak in expression. In order to systematically investigate the role of mRNA decay during germination, 3′ UTRs were examined for enrichment of motifs in transcript subsets that showed peak expression at 3, 12, and 24 HAI. The presence of these predicted motifs (Supplemental Table S6C) and of 12 known RNA stability/instability-associated motifs was compared between the subsets and the “whole genome” set; however, this was somewhat restricted due to the fact that only 3,027 genes have an annotated 3′ UTR in rice (Supplemental Table S6D). Nevertheless, a clear picture emerged, in that four elements were only significantly enriched in 3 HAI, two of which have been associated previously with RNA instability in Arabidopsis (Narsai et al., 2007) and tobacco (Ohme-Takagi et al., 1993; Table I). Interestingly, one sequence element (GAATAA) was associated with stable RNA transcripts (Narsai et al., 2007) and was enriched in transcripts peaking at 12 HAI (Table I). The presence of these putative motifs in the 3′ UTR together with 1-kb upstream motifs suggests that the complex regulation of transcript abundance occurs at the levels of both transcription and degradation during the course of germination.
DISCUSSION
This study provides a comprehensive profile of the transcriptome and metabolites during germination in the monocot model rice. A series of temporal switches in metabolites and transcripts is suggested that results in a reactivation of cellular metabolism to support growth. At the earliest time point analyzed in this study, 1 HAI, there was a greater proportion of the detected metabolites than the detected transcripts changing in abundance relative to the total number of changes observed throughout the time course of this study. These early responses were then followed by the largest change in transcript abundances between 3 and 12 HAI, followed by relatively small changes in transcripts at subsequent time points. In contrast, changes in a large number of metabolites continued up to 48 HAI. This suggests that the early changes in metabolites arise from the activity of preexisting enzymes, as this occurs rapidly, possibly even before the energy-demanding process of translation has been fully activated to synthesize new proteins. However, the later changes in metabolites are more likely driven by transcription and translation, as they occur subsequent to changes in transcript abundance. Furthermore, the changes in transcript abundance that appear transitory in nature, defined in cluster 2, which are enriched in transcription factors but underrepresented in transcripts that encode proteins involved in metabolism, may represent a transition from the dormant state to an active growth state. The peak in transcripts in cluster 2 precedes the increase in abundance of approximately 8,000 transcripts (Fig. 2, cluster 1) but occurs after the decrease in abundance for approximately 8,000 transcripts (Fig. 2, cluster 3). Similar transient peaks in transcript profiles were also observed in a study of germination in Arabidopsis (Nakabayashi et al., 2005), indicating that a multistep transcriptional program appears to be a common theme in seed germination.
A comparison of our rice data with barley seed germination also reveals similarities, with transcripts encoding components involved in sugar, starch, and lipid metabolism being up-regulated, followed by those involved in photorespiration and photosynthesis (Sreenivasulu et al., 2008). Increases in cell wall modification, β-oxidation, tetrapyrrole biosynthesis, amino acid synthesis, energy metabolism genes, and also fermentative components such as ADH were also seen during barley germination (Sreenivasulu et al., 2008).
Upon imbibition, there is an immediate increase in hexose sugars and organic acids that is already statistically significant at 1 HAI (Fig. 2A). It has been previously shown that upon imbibition of rice seeds, there is an immediate increase in water uptake and oxygen consumption in the 1st h, and protein uptake into isolated mitochondria can occur within 30 min of imbibition (Howell et al., 2006, 2007). Thus, this initial stage of metabolic activity likely drives the biogenesis of organelles to produce energy and biosynthetic compounds for subsequent growth. The second burst of changes in metabolites occurs 6 to 12 HAI and involves an increase in many amino acids in addition to maintaining the relatively high levels of many sugars. This increase corresponds to the second phase of increases in oxygen uptake that occurs between 4 and 48 HAI (Howell et al., 2006). These changes in metabolites likely reflect an increase in biosynthetic capacity as well as energy production. At 12 HAI, mitochondria have changed from promitochondrial structures to electron-dense cristae-containing structures, reflecting the increase in various metabolic activities evidenced by the increase in metabolites observed in this study. Thus, the first burst of metabolic activity is likely used to produce energy to build subcellular structures, while the second phase of metabolic activity supports growth. These changes in metabolite and transcript pool sizes are reflected in changes observed in protein abundance. Mitochondria in dry seeds contain large amounts of proteins required to make mitochondria, such as components of the protein import apparatus. However, by 24 HAI, components of the import apparatus have decreased 10-fold or greater in abundance, and components involved in metabolism have increased by at least 10-fold in abundance (Howell et al., 2006).
A number of analyses of transcription factors revealed similarities with previous studies and give insights into the regulatory processes that occur during germination. Transcription factors preferentially expressed in the germinating embryo of barley, such as ARF, AUX/IAA, C2C2-GATA, and C3H-ARFs, were also observed here in rice. This study reveals a greater resolution of these events. Thus, for cluster 3, enriched in the PHD and HSF transcription factor families, and cluster 4, enriched in SET, it can be seen that these transcription factors are present in dry seeds and decrease or remain largely unchanged, respectively (Fig. 7, A and B). In contrast, the transient cluster 2 is enriched in AP2-EREBP and WRKY, while cluster 1 is enriched in the AUX/IAA family and basic helix-loop-helix. Therefore, despite all clusters containing members from several transcription factor families, there is a clear and significant difference in the proportion of families in each cluster, implying an important time-specific regulatory requirement for the expression of these transcription factors. Examination of the transcription factors that peak in expression at 3 HAI (Supplemental Table S4A) reveals that seven of these are AP2-EREBP transcription factors and two are C2H2 zinc finger transcription factors. Previous studies in Arabidopsis have characterized a role for members of the AP2-EREBP family in the regulation of water uptake (Song et al., 2005). Thus, the highly regulated, specific expression pattern of these transcription factors during germination might be related to an important regulatory role of these transcription factors during the water-uptake phases of germination. The enrichment of sequence elements in distinct groups of genes from each cluster (Table I) combined with the transcript abundance data provide a reasonable set of transcription factors that may bind these elements that can be investigated in future studies.
The transcription factors in common with germination and anoxia profiles may also be significant, given that germinating seeds are thought to suffer from oxygen deficit (Bewley, 1997; Borisjuk et al., 2007). These peaked early (3 HAI) during germination (Fig. 8A), at a time when metabolic activity has resumed, energy-demanding processes such as protein synthesis and transcription are active, and yet oxygen diffusion may be limited into the embryo by the endosperm. The expression of transcription factors linked with anoxia is also consistent with an increase in the amount of GABA as early as 1 HAI, which has been proposed to play a role in anoxia tolerance in plants (Fait et al., 2008). Analysis of metabolite changes in Arabidopsis during mild decreases in oxygen concentration revealed that although GABA initially decreased at 0.5 and 2 h, it did increase later at 48 h. However, taking caution in comparing such different systems as rice and Arabidopsis, this may reveal that embryos from seeds with larger endosperms such as rice may be more prone to anoxia and, thus, display alterations in transcriptome and metabolome during germination to avoid detrimental affects.
Approximately 17,000 transcripts are stored in the dry rice embryo during seed development and maturation, compared with approximately 12,000 stored in both barley and Arabidopsis seeds (Nakabayashi et al., 2005; Sreenivasulu et al., 2008). This difference may simply be due to the number of probes represented on each array and the relative sizes of the genomes for each species. In an attempt to uncover insights into the regulatory mechanisms that cause changes in transcript abundance, the enrichment or depletion of sequence elements in the promoters or 3′ UTR was examined. Given that there may be over 2,000 transcription factors and hundreds of stability/instability elements, the prediction of such elements can be hard to interpret. Thus, our analysis was carried out in an attempt to determine if distinct regulatory steps were occurring during germination. Hence, we used stringent criteria with respect to sets of genes used to search for common sequence elements to reveal insights into the regulatory steps that may be occurring during germination. Even with this strict criteria at each time point, with the exception of 1 HAI, a unique enrichment or depletion of groups of elements was displayed, consistent with the combinatorial model of gene regulation and also the fact that a number of transcriptional steps or switches occur during germination. Additionally, the large enrichment of elements associated with RNA instability in the 3′ UTR of transcripts that peaked at 3 HAI indicates that RNA degradation also plays a central role in defining changes in transcript abundance during germination. Although RNA degradation has previously been proposed to “clean out” transcripts that are present in the mature seeds (Rajjou et al., 2004; Nakabayashi et al., 2005), it can be seen in this study that several groups of transcripts decrease in abundance during early germination, as shown in cluster 3 (transcripts that decreased at 0–3 HAI; Fig. 2). However, this study suggests that RNA degradation also plays an important role for specific transcripts that are synthesized after imbibition.
The combination of a specifically timed up-regulation of a suite of specific transcription factors and the degradation of both stored and early-induced mRNAs based on 3′ UTR sequences appear to be key elements in the coordination of at least some groups of transcripts during the early events in rice germination. These events appear to operate in a coordinated fashion with the induction of primary metabolic pathways, the biogenesis of organelles, and the establishment of the full metabolic profile in the germinating rice embryo.
MATERIALS AND METHODS
Rice Growth
Dehulled, sterilized rice seeds (Oryza sativa ‘Amaroo’) were grown under aerobic conditions in the dark at 30°C as described previously (Howell et al., 2006). Embryos were rapidly dissected from the endosperm and snap frozen in liquid nitrogen.
RNA Isolation, cDNA Synthesis, and Quantitative Reverse Transcription-PCR
Total RNA was isolated from rice embryos as described previously (Howell et al., 2006). Three independent RNA preparations were used for each developmental stage/growth condition, and the concentration of RNA was determined spectrophotometrically.
Microarray Analyses
Transcriptomic analysis was performed using Affymetrix GeneChip Rice Genome Arrays (Affymetrix), and three biological replicates were analyzed for each time point. RNA quality was verified using an Agilent Bioanalyzer (Agilent Technologies) and spectrophotometric analysis (NanoDrop ND-1000; NanoDrop Technologies) to determine concentration and the A260-A280 and A260-A230 ratios. Preparation of labeled copy RNA from 2 to 3 μg of total RNA, target hybridization, as well as washing, staining, and scanning of the arrays were carried out exactly as described in the Affymetrix GeneChip Expression Analysis Technical Manual, using the Affymetrix One-Cycle Target Labeling and Control Reagents, an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450, and an Affymetrix GeneChip Scanner 3000 7G at the appropriate steps. Data quality was assessed using GCOS 1.4 (Affymetrix) before CEL files were imported into Avadis 4.3 (Strand Genomics) for further analysis. Raw intensity data were initially normalized using the MAS5 algorithm allowing probe identifications called present to be determined. Only those probe sets that were called present in at least two out of three replicates in at least one time point were included for further analysis. Ambiguous probe sets and bacterial controls were also removed, resulting in a final data set of 24,150-gene set. All microarray data have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under the accession code E-MEXP-1766.
Using the 24,150-gene set, probe intensities were analyzed using the GC-RMA algorithm and log transformed, and differential expression analysis was performed with P value correction (Benjamini and Hochberg, 1995) at the 0.01 level. This allowed the number of transcripts significantly changing to be calculated, which were then visualized on a heat map. For each of the 24,150 transcripts, the maximum expression was assigned a value of 1 and all other expression values were made relative to this, in order to carry out hierarchical clustering. Average linkage hierarchical clustering was carried out, and distinct clusters were uniquely colored for the genome (24,150), mitochondrial, chloroplast, peroxisome, and transcription factor sets. The differential expression analysis was carried out using Avadis 4.3 (Strand Genomics), while the heat maps and hierarchical clustering were all carried out using Partek Genomics suite software, version 6.3 (Partek).
PageMan (Usadel et al., 2006) and MapMan (Thimm et al., 2004; Usadel et al., 2005) analyses were performed using a reduced set of unique probe sets (15,351). Of these, 9,098 were classified into nontrivial MapMan BINS based on the newly available rice mapping file, which was generated by a combination of automated searches in conjunction with minimal curation. In brief, rice protein sequences corresponding to the 15,351 probe sets were obtained from The Institute for Genomic Research (TIGR; version 5.0) and used for searches against five different databases: The Arabidopsis Information Resource (TAIR7) proteins (Swarbreck et al., 2008), SwissProt/Uniprot plant proteins (PPAP; Schneider et al., 2005), Conserved Domain Database (CDD; Marchler-Bauer et al., 2007), Clusters of Orthologous Groups (KOG; Tatusov et al., 2003), and InterProScan (Zdobnov and Apweiler, 2001). The programs used to perform the searches were BLASTP (Altschul et al., 1990) for TAIR7 and PPAP and RPSBLAST (Schaffer et al., 2001) for CDD and KOG. Database hits with bit scores lower than 50 were ignored as not significantly similar. The results of all searches were compiled into one table, and reference mappings of the above-listed databases were then used to assign preliminary MapMan BINcodes to each of the rice proteins. In the next step, the bit scores (in the case of TAIR7, PPAP, CDD, and KOG) for each database hit were recorded and evaluated for each rice protein as a measure of the reliability for the assignment of the protein into certain BINs To finally assign the protein to BINS, the bit scores of all database hits belonging to the same BIN were combined, allowing for multiple assigned BINcodes. In a subsequent step, the resulting BIN assignments were manually compared with the TIGR-based annotation and, in cases of ambiguity, checked against independent information available from gramene.org and the transcription factor database, resulting in more than 300 changes in assignments. Using this file, for both PageMan and MapMan, Wilcoxon rank sum tests with Benjamini-Hochberg false discovery rate control were used to determine statistically significant changes in specific BINS.
Generation of Transcription Factor and Organelle Lists
The transcription factor list was generated using three main sources: DRTF (Gao et al., 2006), RiceTFDB (Riano-Pachon et al., 2007), and Caldana et al. (2007). These lists were compiled, and all unique transcription factors were matched to the 24,150-gene set to generate a list of 1,786 transcription factors. To examine the transcripts encoding mitochondrial, chloroplast, and peroxisomal proteins, it was necessary to generate lists of transcripts known to encode proteins localized to these organelles. First, all large-scale experimental information to date on rice localization was gathered and the transcripts encoding these proteins were automatically assigned to that localization. To date, only a few large-scale localization studies have been carried out, so less than 300 could be assigned in this way. In order to overcome this, all protein sequence information was downloaded for the 24,150 genes, and four primary sources were employed: (1) experimentally shown localization based on protein work (Heazlewood et al., 2003; Howell et al., 2006, 2007; Kleffmann et al., 2007; Schwacke et al., 2007); (2) seven predictor programs: Predotar (Small et al., 2004), Subloc (Chen et al., 2006), TargetP (Emanuelsson et al., 2007), WoLF PSORT (Horton et al., 2007), PTS1 Predictor (Neuberger et al., 2003), PProwler (Boden and Hawkins, 2005; Hawkins and Boden, 2006), and ChloroP (Emanuelsson et al., 1999); (3) Gene Ontology (GO)/keyword information from four databases: Gramene GO cell comp, Affymetrix GO cell comp, TIGR GO cell comp, and TIGR keyword (Yuan et al., 2005); and (4) localization information from orthologous genes in Arabidopsis (Arabidopsis thaliana). When several sources were used in combination, in order for a protein to be assigned to a localization, the cutoffs for these sources were set as follows: (1) for experimentally shown localization, no cutoff was required; (2) at least four out of the seven predictors had to show the same localization; (3) at least two of the four GOs had to be annotated to the same localization; and (4) the transcript had to have at least 50% orthology to the Arabidopsis gene with known localization. Orthology information and GO cellular component information was retrieved from the Gramene database (Jaiswal et al., 2006).
When a transcript was annotated to a particular localization, a “source number” was assigned to represent the source used to determine this localization. The source numbers were representative as follows: 1, localization based on experimental evidence; 2, two of the four primary sources agreed on localization (i.e. cutoffs were met in at least two primary sources); 3, three out of four primary sources agreed on localization; and 4, all four of the primary sources agreed on localization. For some transcripts, there was only information from one primary source; therefore, the cutoffs for some sources were raised to maintain stringency. Thus, transcripts with a source number between 7 and 9 represent transcripts for which there was only information from one of the four primary sources with numbers assigned as follows: 7, these transcripts had >70% identity with the orthologous gene in Arabidopsis with known localization (for peroxisomes, this cutoff was allowed to be lowered to >50%, as the prediction programs and other sources did not provide equivalent coverage for detecting peroxisomal genes); 8, for these transcripts, three of the four GO-related localization sources were annotated to be in the same localization (for peroxisomes, two out of four was sufficient); 9, at least four of the seven predictors agreed on localization. For peroxisomes, only one predictor was sufficient, as most of the prediction programs did not even have peroxisome as a choice of localization; therefore, the PTS1 Predictor default cutoff was deemed to be a sufficiently stringent. The source number 10 shows that none of the sources produced any conclusive organelle localization information, even at the lowered standards, while a source number of 11 indicates that one or more of the cutoff criteria were met but the localization based on these methods was conflicting between sources.
Functional Annotation and Statistical Analysis
For each probe set, the GO annotations and transcript assignment were as retrieved from Affymetrix. The National Science Foundation rice microarray database was used to match each Affymetrix probe identifier to a National Science Foundation accession identifier and to a TIGR locus identifier. These TIGR locus identifiers were then entered into the TIGR rice database, and the putative function of the encoded proteins was derived (Yuan et al., 2005). The Rice Annotation Project (RAP) database was also used to gather information about function, including the RAP description and RAP GO description. In order to gather this information, the files available from the RAP database were first used to convert each TIGR locus identifier to a RAP Os identifier. Lastly, in order to categorize the transcripts based on the FUNctional CATalogue (FUNCAT) of the encoded protein, the Australian National University genebins database was used for the whole genome set. Two FUNCATs were independently added: transcription factors, which was formed as a separate category based on DRTF (Gao et al., 2006), RiceTFDB (Riano-Pachon et al., 2007), and Caldana et al. (2007); and kinases, which was based on the rice kinase database (Dardick et al., 2007). For the organelle lists, the broad FUNCATs (Australian National University), the FUNCATs based on previously published data (Heazlewood et al., 2003), the FUNCAT of the orthologous gene in Arabidopsis, and manual annotation were used so that as many of the organelle genes as possible could be assigned a function. In order to compare the difference between the percentage of genes in a given FUNCAT within the genome set with the percentage of genes in that FUNCAT in a given cluster, z-score analysis was carried out to determine the significance of the difference between the two proportions, given that we know the sample sizes, frequency, and percentages for each set:
 |
The z-scores were then matched to the cumulative standard normal table, and the P values were determined.
Public Rice Microarray Data Analysis and Comparison
In order to examine transcript abundance changes across different tissues under different conditions and compare these with the germination transcript abundance profiles generated from this study, rice array data were retrieved from the Gene Expression Omnibus within the National Center for Biotechnology Information database. All data were MAS5.0 normalized and normalized against average ubiquitin expression for that array. These normalized array data were then compiled together, and for each probe set, the maximum expression was set to 1.0 with all other data relative to this. This normalization allowed cross-comparison of arrays from all of the different studies at once. The arrays analyzed included all of the arrays from this study, together with publicly available rice genome arrays carried out from different tissues/conditions, including 7-d-old seedlings that were untreated, drought stressed, salt stressed, or cold stressed (GSE6901; Jain et al., 2007); seeds collected at 5 d following pollination, 10-d-old embryos, 10-d-old endosperms, seedling roots, seedling shoots, unpollinated stigmas (at antithesis), ovaries (at antithesis), mature anthers, and suspension cells (GSE7951; Li et al., 2007); aerobically grown coleoptiles (4 d) and anoxically grown coleoptiles (4 d; GSE6908; Lasanthi-Kudahettige et al., 2007); crowns and growing points under salt stress and control conditions in sensitive and tolerant mutants in subspecies indica and japonica (GSE4438; Walia et al., 2007); crowns and growing points under control and salt stress conditions in subspecies indica and japonica (GDS1383; Walia et al., 2005); and leaves following biotic stress and control treatments (GSE7256; Ribot et al., 2008).
Promoter Motif Analysis
Following expression analysis, distinct groups of transcripts appeared that showed peak expression at single specific time points within the time course. In order to study these coexpressed transcripts more closely, all 1-kb upstream regions of the 24,150 transcripts were retrieved, and these upstream regions were examined for putative cis-acting elements. Programs designed to detect sequence elements generally have limits of less than 80 input sequences; thus, the list was distilled to uncover sequence elements that may be central to the regulatory processes that cause the changes in transcriptome observed. A “peak” was defined as a probe set having an expression value of 1.0 at that specific time point with expression levels of less than 0.5 at all other time points. Three main cis-element databases were used for this analysis. The first was the Rice Cis-Element Search database (Doi et al., 2008), which was used under default settings and searched for enrichment of known plant cis elements in the 1-kb upstream region. The second database used was the MEME Web server (Bailey et al., 2006), which was used under default settings with the length of the motif set to 6 to 8 bp and the number of motifs to find set to five (instead of the default of three). The MEME database could not process the large data sets, including the plastid and transcription factor 24-h peak subsets, so no output was generated for these. The third database used was the Regulatory Sequence Alignment Tool (Thomas-Chollier et al., 2008), which was used under default settings, with the only exception being that Markov modeling was selected for background calculation, as Oryza sativa was not available as a choice for the background model. The outputs from all of these databases are shown in Supplemental Table S6B. The lists of motifs from Supplemental Table S6B were then filtered only to include motifs present in more than 70% of all input sequences (Supplemental Table S6C), and the presence of these motifs was then examined in the whole genome and the genome “peaking” subsets, where a peak is as defined above.
3′ UTR Sequence Analysis
The full genome 3′ UTR and 5′ UTR sequences are available from TIGR. This was downloaded and filtered to retain only the 3′ UTRs. However, this only added up to 3,027 UTRs available for the “whole genome.” Taking this small number into consideration, it was not feasible to look at the organelle-specific and transcription factor peaking subsets analyzed for the promoter regions, as these lists were too small. Thus, for the 3′ UTR, the genes peaking in expression at 0, 1, 3, 12, and 24 HAI in the entire genome set were analyzed; however, there were still too few in the 0- and 1-HAI peaking subsets, so these could not be analyzed (Table I). In order to look at the enrichment of motifs in an objective manner, only the MEME Web server was used, as we were not searching for known regulatory elements. The settings were set to search for five motifs that are 6 to 8 bp (default) in each of the subsets, and the outputs are shown at the bottom of Supplemental Table S6D. It is important to note that setting the output to be five motifs can result in false present calls for motifs that are not significant when the input list is small; therefore, only the significantly enriched motifs (present in 60%−70% of all input sequences) were included for further analysis (Supplemental Table S6, C and D). In addition to these putative predicted motifs, 12 motifs known to be associated with RNA stability/instability were examined for their presence in the genome (Table I; Supplemental Table S6D). Ten of these were motifs predicted to be associated with stability/instability of mRNA (Narsai et al. 2007), and two elements had previously been shown to be associated with RNA stability/instability (Newman et al., 1993; Ohme-Takagi et al., 1993).
Metabolomic Analysis
Data for the 126 nonredundant metabolites were analyzed by two-way differential comparisons to determine fold changes and associated P values, and the number of metabolites significantly changing were also visualized by heat map. The heat map showing the number of significantly changing metabolites was generated using Partek Genomics suite software, version 6.3.
Extraction and Derivatization of Metabolites for Gas Chromatography-Mass Spectrometry Analysis
Metabolites were extracted and derivatized using a method modified from that of Roessner-Tunali et al. (2003). To each tube containing 20 to 40 mg of frozen tissue powder was added 300 μL of cold (−20°C) Metabolite Extraction Medium (85% [v/v] HPLC-grade methanol [Sigma], 15% [v/v] untreated MilliQ water, and 100 ng μL−1 ribitol), and tubes were vortexed briefly and shaken at 1,400 rpm for 15 min at 70°C. Tubes were then centrifuged at 13,000g for 3 min to pellet insoluble material, and supernatant was reextracted with chloroform. Aliquots (100 μL) of the methanol fraction were dried under vacuum in 1.5-mL microfuge tubes. Dried extracts were methoximated by adding 20 μL of a 20 mg mL−1 solution of methoxyamine hydrochloride in anhydrous pyridine (Sigma) and incubating at 30°C for 90 min with shaking at 1,400 rpm. For trimethylsilylation, 30 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma) was transferred to each tube, and tubes were incubated at 37°C for 30 min with 1,400 rpm shaking. Ten microliters of an n-alkane retention index calibration mixture (0.29% [v/v] n-dodecane, 0.29% [v/v] n-pentadecane, 0.29% [w/v] n-nonadecane, 0.29% [w/v] n-docosane, 0.29% [w/v] n-octacosane, 0.29% [w/v] n-dotracontane, and 0.29% [w/v] n-hexatriacontane dissolved in anhydrous pyridine) was then added to each tube, and reaction mixtures were vortexed and transferred to amber gas chromatography-mass spectrometry (GC-MS) vials with low-volume inserts and screw-top seals (Agilent Technologies) and allowed to rest for 4 h prior to beginning GC-MS analysis.
GC-MS Instrumental Analysis
Derivatized metabolite samples were analyzed on an Agilent GC/MSD system composed of an Agilent GC 6890N gas chromatograph (Agilent Technologies) fitted with a 7683B Automatic Liquid Sampler (Agilent Technologies) and 5975B Inert MSD quadrupole MS detector (Agilent Technologies). The gas chromatograph was fitted with a 0.25-mm (i.d.), 0.25-μm film thickness, 30-m Varian FactorFour VF-5ms capillary column with 10 m integrated guard column (Varian; product no. CP9013). GC-MS run conditions were essentially as described for GC-quadrupole-MS metabolite profiling on the Golm Metabolome Database Web site (http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/analytic/gmd_meth.html; Kopka et al., 2005). Samples were injected into the split/splitless injector operating in splitless mode with an injection volume of 1 μL, purge flow of 50 mL min−1, purge time of 1 min, and a constant inlet temperature of 300°C. Helium carrier gas flow rate was held constant at 1 mL min−1. The GC column oven was held at the initial temperature of 70°C for 1 min before being increased to 76°C at 1°C min−1 and then to 325°C at 6°C min−1 before being held at 325°C for 10 min. Total run time was 58.5 min. Transfer line temperature was 300°C. MS source temperature was 230°C. Quadrupole temperature was 150°C. Electron-impact ionization energy was 70 eV, and the MS detector was operated in full scan mode in the mass-to-charge ratio range 40 to 600 with a scan rate of 2.6 Hz. The MSD was pretuned against perfluorotributylamine mass calibrant using the “atune.u” autotune method provided with the Agilent GC/MSD Productivity ChemStation software (version D.02.00 SP1; Agilent Technologies; product no. G1701DA).
GC-MS Data Analysis
Raw GC-MS data files in the proprietary ChemStation (.D) format were exported to generic NetCDF/AIA (.CDF) format with ChemStation GC/MSD Data Analysis software (Agilent Technologies). The NetCDF files produced were then processed using in-house MetaMiner software (A. Carroll and A.H. Millar, unpublished data) to carry out all peak detection, quantification, library matching, normalization, statistical analysis, and data visualization. Raw data processing in MetaMiner consisted of the following steps: retrieval of all extracted ion chromatograms (EICs), detection and integration of peaks in EICs, calculation of internally calibrated retention indices for all extracted peaks, matching of carefully selected analyte-specific EIC peaks to analytes in a custom mass spectral-retention index (MSRI) library of known and unknown metabolite derivatives (retention index error < 3 retention index units; Wagner et al., 2003; Schauer et al., 2005), and normalization of matched peak areas to the peak area of the internal standard, ribitol, and to fresh tissue weight of extracted samples. The MSRI library was constructed using publicly available AMDIS software (version 2.65) to extract MSRI information for authentic standard derivatives from standard runs and MSRI information for unknown analytes from representative analyses of complex biological extracts. In a few cases, certain analyte peaks were assigned a putatively known annotation based on matching to the Q_MSRI_ID MSRI library (version 2004-03-01) available from the Golm Metabolome Database (Kopka et al., 2005). In these cases, positive identification required a “weighted” mass spectral match score of greater than 90 and a retention index discrepancy of less than 2%. Unknown metabolite derivative peaks that could not be putatively identified by comparison with authentic standards or by matching against the Q_MSRI_ID library were annotated with a simple generic identifier with the syntax USH: name, match_score, where USH stands for “unknown spectral homolog,” name is the abbreviated name of top NIST02 mass spectral library match, and match_score is the “simple” match score reported by AMDIS. Artifact peaks and common contaminants were identified by analysis of negative control samples prepared in the same manner as biological samples but without the inclusion of tissue. Signals corresponding to these artifacts were not used in biological interpretation. Automatic statistical analysis of processed data was carried out by calculating, for each set of biological replicates, the mean signal intensity for each metabolite, and then, for each metabolite, dividing the mean signal in treated sample sets by the mean signal in control sample sets to calculate fold difference and testing the statistical significance (P < 0.05) of this difference by Student's t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Overview of the changes in transcripts and metabolites during germination in rice.
Supplemental Figure S2. Two-way comparison of 0 versus 3 HAI and 3 versus 12 HAI using MapMan to visualize an overview of regulation.
Supplemental Figure S3. Two-way comparison of 0 versus 3 HAI and 3 versus 12 HAI using MapMan to visualize an overview of metabolism.
Supplemental Figure S4. Functional categorization of the transcripts grouped into each cluster.
Supplemental Figure S5. All 74 transcripts encoding proteins predicted to be peroxisomal were hierarchically clustered.
Supplemental Figure S6. Phylogenetic analysis of the homeobox (HB) transcription factor family (A) and the zinc finger homeodomain (zf-HD) transcription factor family (B) in rice and Arabidopsis.
Supplemental Table S1. All 24,150 expressed genes and the calculated fold changes between combinations of time points.
Supplemental Table S2. Averaged raw metabolite abundance data with standard errors for the 256 detected metabolites.
Supplemental Table S3. FUNCAT information from Figure 3.
Supplemental Table S4. The transcript abundance profiles of all 1,786 transcription factors were hierarchically clustered and the order of the transcripts following clustering is shown with functional information.
Supplemental Table S5. Transcripts encoding proteins predicted/experimentally shown to be located in plastids, mitochondria, or peroxisomes.
Supplemental Table S6. Sequence analysis of all rice 1-kb upstream sequences and known 3′ UTR sequences.
Supplementary Material
Acknowledgments
We thank Ian Castleden from the Centre for Computational Systems Biology for help with the multiple localization predictions and sequence matching.
This work was supported by the Australian Research Council Centre of Excellence (grant no. CEO561495) and an Australian Research Council Australian Professorial Fellowship to A.H.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James Whelan (seamus@cyllene.uwa.edu.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34 W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Fung P, Chow TF, Foong JA, Provart NJ, Cutler SR (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B Methodological 57 289–300 [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden M, Hawkins J (2005) Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21 2279–2286 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Macherel D, Benamar A, Wobus U, Rolletschek H (2007) Low oxygen sensing and balancing in plant seeds: a role for nitric oxide. New Phytol 176 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ (2007) Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol 143 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Huang N, Sun Z (2006) SubLoc: a server/client suite for protein subcellular location based on SOAP. Bioinformatics 22 376–377 [DOI] [PubMed] [Google Scholar]
- Conte MG, Gaillard S, Lanau N, Rouard M, Perin C (2008) GreenPhylDB: a database for plant comparative genomics. Nucleic Acids Res 36 D991–D998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick C, Chen J, Richter T, Ouyang S, Ronald P (2007) The rice kinase database: a phylogenomic database for the rice kinome. Plant Physiol 143 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Hosaka A, Nagata T, Satoh K, Suzuki K, Mauleon R, Mendoza MJ, Bruskiewich R, Kikuchi S (2008) Development of a novel data mining tool to find cis-elements in rice gene promoter regions. BMC Plant Biol 8 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L, Waters L (1965) Long-lived messenger RNA: evidence from cotton seed germination. Science 147 410–412 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols 2 953–971 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13 14–19 [DOI] [PubMed] [Google Scholar]
- Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, Gu X, Wei L, Luo J (2006) DRTF: a database of rice transcription factors. Bioinformatics 22 1286–1287 [DOI] [PubMed] [Google Scholar]
- Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J (2008) Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genomics 35 105–118 [DOI] [PubMed] [Google Scholar]
- Hawkins J, Boden M (2006) Detecting and sorting targeting peptides with neural networks and support vector machines. J Bioinform Comput Biol 4 1–18 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Howell KA, Whelan J, Millar AH (2003) Towards an analysis of the rice mitochondrial proteome. Plant Physiol 132 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ (2008. a) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2008. b) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13 7–13 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35 W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Cheng K, Murcha MW, Jenkin LE, Millar AH, Whelan J (2007) Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem 282 15619–15631 [DOI] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J (2006) Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Mol Biol 60 201–223 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, Ratnapu K, et al (2006) Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res 34 D717–D723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol 143 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21 1635–1638 [DOI] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarova G, Zeng Y, Kermode AR (2002) Cloning and expression of an ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene homologue of yellow-cedar (Chamaecyparis nootkatensis). J Exp Bot 53 1219–1221 [DOI] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y (2007) Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 144 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35 D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F (2003) Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J Mol Biol 328 567–579 [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Taylor CB, Newman TC, Green PJ (1993) The effect of sequences with high AU content on mRNA stability in tobacco. Proc Natl Acad Sci USA 90 11811–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J 410 621–629 [DOI] [PubMed] [Google Scholar]
- Prohl C, Pelzer W, Diekert K, Kmita H, Bedekovics T, Kispal G, Lill R (2001) The yeast mitochondrial carrier Leu5p and its human homologue Graves' disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol 21 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314 1041–1052 [DOI] [PubMed] [Google Scholar]
- Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics 8 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Notteghem JL, Lebrun MH, Morel JB (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea. J Plant Physiol 165 114–124 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schneider M, Bairoch A, Wu CH, Apweiler R (2005) Plant protein annotation in the UniProt Knowledgebase. Plant Physiol 138 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Fischer K, Ketelsen B, Krupinska K, Krause K (2007) Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol Genet Genomics 277 631–646 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J (2008) RSAT: regulatory sequence analysis tools. Nucleic Acids Res 36 W119–W127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62 887–900 [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, et al (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Zeng L, Ismail AM, Condamine P, Close TJ (2007) Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol Biol 63 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson L, Henry RJ (2005) Microarray analysis of gene expression in germinating barley embryos (Hordeum vulgare L.). Funct Integr Genomics 5 155–162 [DOI] [PubMed] [Google Scholar]
- Wilson ID, Barker GL, Lu C, Coghill JA, Beswick RW, Lenton JR, Edwards KJ (2005) Alteration of the embryo transcriptome of hexaploid winter wheat (Triticum aestivum cv. Mercia) during maturation and germination. Funct Integr Genomics 5 144–154 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Wilson JR, Gamblin SJ (2003) SET domains and histone methylation. Curr Opin Struct Biol 13 699–705 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol 138 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17 847–848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.