Abstract
Nitrate releases seed dormancy in Arabidopsis (Arabidopsis thaliana) Columbia accession seeds in part by reducing abscisic acid (ABA) levels. Nitrate led to lower levels of ABA in imbibed seeds when included in the germination medium (exogenous nitrate). Nitrate also reduced ABA levels in dry seeds when provided to the mother plant during seed development (endogenous nitrate). Transcript profiling of imbibed seeds treated with or without nitrate revealed that exogenous nitrate led to a higher expression of nitrate-responsive genes, whereas endogenous nitrate led to a profile similar to that of stratified or after-ripened seeds. Profiling experiments indicated that the expression of the ABA catabolic gene CYP707A2 was regulated by exogenous nitrate. The cyp707a2-1 mutant failed to reduce seed ABA levels in response to both endogenous and exogenous nitrate. In contrast, both endogenous and exogenous nitrate reduced ABA levels of the wild-type and cyp707a1-1 mutant seeds. The CYP707A2 mRNA levels in developing siliques were positively correlated with different nitrate doses applied to the mother plants. This was consistent with a role of the CYP707A2 gene in controlling seed ABA levels in response to endogenous nitrate. The cyp707a2-1 mutant was less sensitive to exogenous nitrate for breaking seed dormancy. Altogether, our data underline the central role of the CYP707A2 gene in the nitrate-mediated control of ABA levels during seed development and germination.
Plants adapt to their environment by controlling their development in response to different external cues such as light, photoperiod, temperature, and nutrient availability. The seed phase represents a crucial stage in plant development in which plant survival is determined in part by seed germination at a proper time when external conditions become favorable. Seed dormancy, a temporary quiescent state that is observed in seeds from many plant species, prevents untimely germination and ensures plant survival by adjusting vegetative development to seasonal changes in the environment (Donohue et al., 2005). Many external factors are known to reduce or break seed dormancy after dispersal (Finch-Savage and Leubner-Metzger, 2006): imbibition at low temperature, the presence of nitrogenous compounds such as nitric oxide (NO) or nitrate, and after-ripening (a period of dry storage of mature seeds). Not surprisingly, seed primary dormancy, which is observed in freshly harvested seeds, depends also on the environmental conditions during seed maturation on the mother plants. Temperature, photoperiod, and light quality experienced by the mother plant thus constitute important environmental cues affecting the dormancy of the produced seeds (McCullough and Shropshire, 1970; Hayes and Klein, 1974; Munir et al., 2001; Donohue et al., 2007).
Genetic and physiological studies have underlined the importance of two hormones, abscisic acid (ABA) and GAs, in seed dormancy and germination (Bentsink and Koornneef, 2002). ABA is a key hormone promoting dormancy. Thus, a number of mutants affected in ABA synthesis or response display altered dormancy (Finkelstein et al., 2002; Gubler et al., 2005). More recently, the Arabidopsis (Arabidopsis thaliana) CYP707A1 and CYP707A2 genes, which belong to a small multigene family involved in ABA catabolism, were shown to control seed dormancy: mutants affected in either of these genes were more dormant than wild-type seeds and accumulated high levels of ABA in seeds. Yet spatial and temporal expression profiles of these genes during seed development and germination were different, suggesting distinct roles for the CYP707A1 and CYP707A2 genes (Okamoto et al., 2006). Indeed, the CYP707A2 gene was shown to play a major role in ABA degradation early during seed imbibition. Its mRNAs accumulated highly in dry seeds and were induced early (6 h) upon imbibition, whereas CYP707A1 gene expression peaked in siliques and increased moderately at the later stages of seed imbibition (Kushiro et al., 2004). GAs, on the other hand, promote germination (Yamaguchi and Kamiya, 2002), and GA-deficient mutants fail to germinate (Ross et al., 1997). The importance of the ABA/GA balance for germination is supported by the fact that the reduced dormancy of some of the ABA mutants was linked with a lowered requirement for GA for germination (Koornneef et al., 2002). At the molecular level, the ABA/GA balance is in part determined by the antagonistic control of ABA and GA on each other through their reciprocal regulation of the transcription of their metabolic genes (Seo et al., 2006; Oh et al., 2007).
Transcriptome approaches in Arabidopsis have further sustained the importance of ABA and GA in controlling dormancy cycling. Indeed, genes with ABA-responsive elements and genes involved in ABA synthesis were more highly expressed in dormant (D) seeds than in nondormant (ND) seeds (Cadman et al., 2006; Finch-Savage et al., 2007). Conversely, GA biosynthesis genes were more highly expressed in ND seeds than in D seeds (Cadman et al., 2006; Finch-Savage et al., 2007). These transcriptome studies further highlighted the distinct gene expression programs of ND and D seeds with a higher representation of protein synthesis-related genes in ND seeds and stress-related genes in D seeds (Cadman et al., 2006). Different dormancy-releasing treatments led to similar profiles, sustaining the idea that the seeds displayed a similar physiological dormancy state irrespective of the difference in environmental cues (Finch-Savage et al., 2007).
Physiological and genetic analyses have also helped uncover important genes involved in the control of seed germination and dormancy by cold or light experienced during seed maturation or imbibition (Oh et al., 2004; Penfield et al., 2005; Donohue et al., 2008). Thus, the AtGA3ox1 gene was shown to mediate the increase of active GAs in response to low temperature (Yamauchi et al., 2004). Likewise, the basic helix-loop-helix SPATULA and PHYTOCHROME-INTERACTING FACTOR3-LIKE5 transcription factors were reported to be involved in seed germination responses, respectively, to light and to light and low temperature by affecting the expression of GA- or ABA-related genes (Oh et al., 2004, 2007; Penfield et al., 2005).
Nitrate has long been known to release seed dormancy (Hilhorst and Karssen, 1989; Alboresi et al., 2005; Bethke et al., 2006b), possibly by affecting seed light requirement (Batak et al., 2002) or ABA levels (Ali-Rachedi et al., 2004). When added in the germination medium, nitrate led to higher germination percentages of Arabidopsis D seeds. This effect (exogenous effect of nitrate) was likely due to a signaling effect of nitrate per se, since Gln, another nitrogen source, did not relieve dormancy (Alboresi et al., 2005). More recently, nitrate, nitrite, and cyanide were shown to relieve seed dormancy in Arabidopsis likely via the generation of NO, since cPTIO, a NO scavenger, prevented dormancy removal by these nitrogenous compounds (Bethke et al., 2006b).
In addition to this exogenous effect of nitrate, a maternal effect of nitrate on seed dormancy was evidenced. Indeed, mother plant nitrate nutrition was shown to affect seed dormancy (Alboresi et al., 2005). Higher nitrate nutrition (50 mm) during seed maturation led to the production of less dormant seeds than those produced under standard nitrate nutrition (10 mm). The latter seeds, however, were less dormant than those obtained under low-nitrate feeding of mother plants (3 mm). Interestingly, a genotype deficient in nitrate assimilation, the nia1 nia2 mutant (Wilkinson and Crawford, 1993), accumulated nitrate under standard nitrate nutrition and produced seeds that were less dormant than wild-type seeds obtained in the same conditions. Altogether, these data suggested that the depth of seed dormancy was inversely correlated to seed nitrate content (referred to as “endogenous nitrate”), whether higher nitrate content was due to mother plant feeding or to a mutation leading to nitrate accumulation (Alboresi et al., 2005). At the molecular level, however, little is known about the players mediating nitrate control of seed dormancy. Physiological analyses of Arabidopsis plants defective in the nitrate dual-affinity NRT1.1 transporter gene and more recently in the nitrate transporter NRT2.7 gene revealed a possible involvement of these transporters in nitrate signaling or accumulation in seeds (Alboresi et al., 2005; Chopin et al., 2007).
This work analyzes the effect of both exogenous and endogenous nitrate on seed dormancy by assessing ABA levels and performing transcriptome analyses in nitrate-treated seeds. We show here that effects of both exogenous and endogenous nitrate were correlated with lower accumulation of ABA in nitrate-treated seeds and higher expression of the ABA catabolic gene CYP707A2. Physiological analyses show that the CYP707A2 gene but not the CYP707A1 gene was specifically involved in mediating changes in ABA levels and seed dormancy in response to exogenous nitrate. Expression of the CYP707A2 gene during seed development was consistent with its involvement in controlling seed ABA levels by endogenous nitrate. Altogether, our data underline the central role of the CYP707A2 gene in the control by exogenous and possibly endogenous nitrate of seed ABA levels.
RESULTS
ABA Levels Were Reduced in Nitrate-Treated Seeds
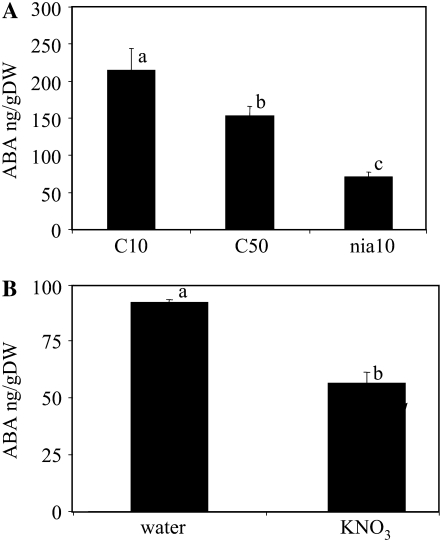
Our previous results showed that both exogenous nitrate and endogenous nitrate released seed dormancy (Alboresi et al., 2005). Indeed, freshly harvested Columbia accession (Col-0) wild-type seeds obtained under standard nitrate nutrition (10 mm nitrate) did not germinate in water but did in the presence of 10 mm nitrate (exogenous nitrate treatment). Also, higher nitrate nutrition (50 mm) of wild-type mother plants or a genotype (nia1 nia2, deficient in nitrate reductase activity) accumulating nitrate under standard nutrition led to the production of ND seeds (endogenous nitrate “treatments”). Since ABA is important for controlling seed dormancy, we measured ABA levels in the ND nitrate-treated and the D nontreated seeds by HPLC followed by immunodetection. To test the effect of endogenous nitrate, ABA content was measured in freshly harvested wild-type seeds produced under standard nutrition (C10 D seeds) or high-nitrate nutrition (50 mm nitrate feeding; C50 ND seeds) and in nia1 nia2 seeds produced under standard nutrition (nia10 ND seeds). Both C50 and nia10 dry seeds accumulated significantly lower ABA levels than C10 dry seeds (P < 0.05; Fig. 1A), consistent with the lower dormancy of seeds treated with endogenous nitrate. To analyze the effect of exogenous nitrate, ABA levels were determined in C10 seeds imbibed for 20 h in the presence or not of nitrate. Imbibition of C10 seeds in the presence of 10 mm nitrate (exogenous nitrate) resulted in significantly lower ABA levels (P < 0.05) compared with control seeds imbibed in water (Fig. 1B). Altogether, these data showed that the lower dormancy of seeds treated with exogenous or endogenous nitrate was correlated with lower ABA levels.
Figure 1.
ABA contents of nitrate-treated seeds. A, ABA contents of seeds treated with endogenous nitrate. ABA content was determined in freshly harvested dry seeds from wild-type Col-0 plants grown with 10 mm nitrate (C10 control), with 50 mm nitrate (C50), and in the nia1 nia2 mutant seeds obtained with 10 mm nitrate nutrition (nia10). B, ABA contents of seeds treated with exogenous nitrate. Freshly harvested seeds from Col-0 plants cultivated with 10 mm nitrate were imbibed for 20 h in water or 10 mm KNO3, and ABA content was determined by HPLC followed by immunodetection. Error bars indicate se (n = 5–7). Letters indicate the different groups displaying significantly different ABA levels (P < 0.05) after ANOVA analysis followed by Fisher's lsd test. DW, Dry weight.
Transcriptome Analysis Revealed Distinct Profiles for Seeds Treated with Exogenous or Endogenous Nitrate
Transcriptome analyses were performed to compare the transcript profiles of nitrate-treated and nontreated seeds, either dry seeds or imbibed seeds, using the CATMA arrays (Crowe et al., 2003; Hilson et al., 2004). We chose a short time point for seed imbibition so that no macroscopic difference is observed between D and ND seeds. A preliminary time-course study of ABA levels during C10 seed imbibition (0–12 h) showed that 6 h of imbibition was sufficient for seed ABA levels to decrease to the basal level and that this level was higher in water-imbibed than in nitrate-imbibed seeds (data not shown). Therefore, we chose 6 h of imbibition as the time point for transcriptome studies on imbibed seeds. To determine the effect of endogenous nitrate, mRNA profiles were compared between ND (C50 or nia10) and D (C10) seeds either as dry seeds (C50ds versus C10ds, niads versus C10ds experiments) or as water-imbibed seeds (C50w versus C10w, niaw versus C10w). Exogenous nitrate was studied by comparing C10 seeds imbibed in 10 mm nitrate (C10NO3, ND) and C10 seeds imbibed in water (C10w, D; C10NO3 versus C10w experiment). Altogether, five comparisons were made corresponding to the different nitrate treatments on the different seed batches (Table I).
Table I.
Comparisons performed for transcriptome experiments
RNAs extracted from control and test samples were used for hybridizations with the CATMA arrays. C10 and C50, Col-0 wild-type freshly harvested seeds obtained, respectively, with 10 or 50 mm mother plant nitrate nutrition; nia10, freshly harvested nia1 nia2 seeds obtained with 10 mm mother plant nitrate nutrition; w, seeds imbibed for 6 h in water; NO3, seeds imbibed for 6 h in 10 mm KNO3; ds, dry seeds.
| Studied Process | Experiment | Compared Samples
|
|
|---|---|---|---|
| Test Sample (ND Seeds) | Control Sample (D Seeds) | ||
| Effect of exogenous nitrate on seed dormancy | C10NO3 versus C10w | C10NO3 | C10w |
| Effect of endogenous nitrate on seed dormancy | C50w versus C10w | C50w | C10w |
| C50ds versus C10ds | C50ds | C10ds | |
| nia10w versus C10w | nia10w | C10w | |
| nia10ds versus C10ds | nia10ds | C10ds | |
Two independent biological replicates were performed using RNAs extracted from seeds obtained in two independent cultures. Genes were considered as differentially expressed when in both replicates they showed a Bonferroni P value of <0.05 (see “Materials and Methods”) with a log2 ratio of either ≥0.7 or ≤−0.7. Altogether, 281 genes were found to be differentially expressed at least in one nitrate treatment compared with untreated control seeds. For each of the five comparisons performed, only a few genes (28–119 genes, depending on the comparisons performed) were thus identified as differentially expressed in nitrate-treated versus nontreated seeds (Fig. 2; Supplemental Tables S1–S5). There were no genes common to the three experiments studying the effect of endogenous or exogenous nitrate in imbibed seeds (C50w versus C10w, niaw versus C10w, and C10NO3 versus C10w experiments), and few genes (4–17) were common to two of these three experiments (Supplemental Fig. S1). Thus, the profiles of the different nitrate treatments led to distinct seed mRNA profiles. Quantitative reverse transcription (qRT)-PCR on a small set of genes (Supplemental Fig. S2) confirmed on the whole the expression profiles of these genes but suggested that the low number of common genes could be partly due to the stringent statistical tests used to identify differentially expressed genes in the microarray experiments. A vast majority of the genes that were differentially expressed in more than one comparison (62 out of 64 genes) presented a similar profile (either always repressed or always induced) in all of the comparisons where they were differentially expressed (Supplemental Fig. S1; Supplemental Tables S6–S8).
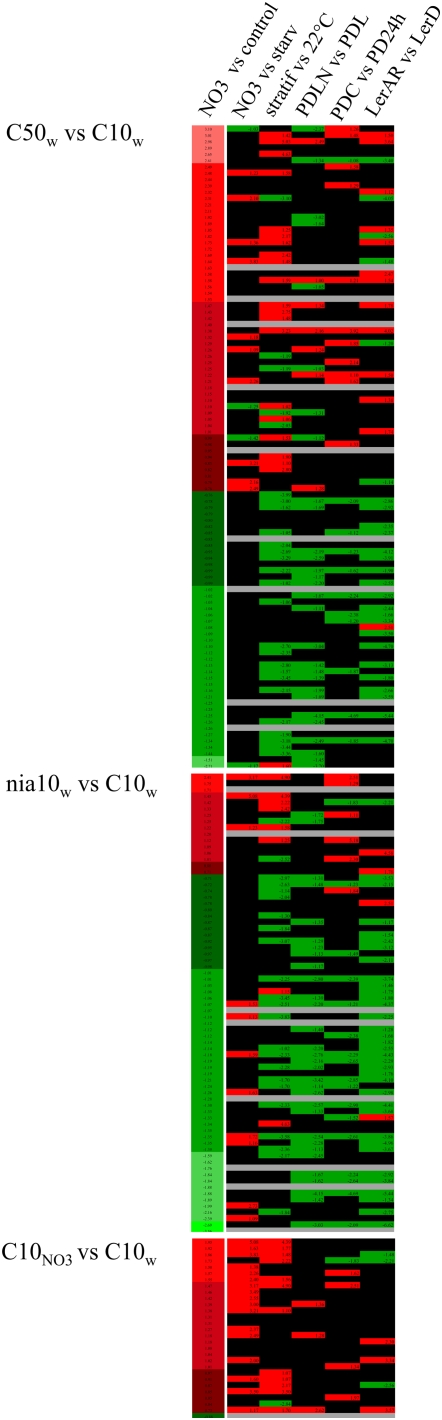
Figure 2.
Profiles of genes differentially expressed (P < 0.05) in imbibed nitrate-treated seeds versus control seeds and their profiles in nitrate and seed transcriptomes. The different transcriptome experiments with imbibed nitrate-treated seeds versus control imbibed C10 seeds are indicated on the left as well as the log2 ratios of gene expression in tested versus control conditions (first column). Red shading indicates overexpression, green shading indicates underexpression in tested versus control samples, and black shading indicates no differential expression. Profiles are compared with those in published array data (second to sixth columns): nitrate provision to starved plants (Scheible et al., 2004; NO3 versus starv); comparison of Ler seeds imbibed at 4°C and 22°C imbibed seeds (Yamauchi et al., 2004; stratif versus 22°C); nitrate-treated primary dormant Cvi seeds after-ripened for 91 d and then imbibed for 24 h in the light compared with the same seeds imbibed in the light without nitrate (Finch-Savage et al., 2007; PDLN versus PDL); cold-treated primary dormant Cvi seeds after-ripened for 117 d compared with 20°C imbibed primary dormant Cvi seeds in the dark (Finch-Savage et al., 2007; PDC versus PD24h); and after-ripened Ler seeds imbibed for 24 h compared with freshly harvested Ler seeds imbibed for 24 h (Carrera et al., 2007; Ler AR versus Ler D). In all cases, log2 ratios of expression are indicated. Gray shading indicates genes absent on the Affymetrix chips. Gene identifiers and functions as well as the values of the log2 ratios in the different experiments are provided in Supplemental Tables S1 to S3.
The transcriptomes of D and ND imbibed seeds can be distinguished by the functional categories associated with their differentially expressed genes (Cadman et al., 2006). In addition, genes up-regulated in after-ripened (AR) seeds belong to different functional categories (as redefined for processes linked with dormancy, germination, and embryo maturation) than genes up-regulated in freshly harvested seeds (Carrera et al., 2007, 2008), leading to signatures typical of AR or freshly harvested seeds. In our case, however, the small number of differentially expressed genes between nitrate-treated and nontreated seeds hindered analysis by functional classification. Therefore, to get a better image of the genes differentially expressed in the various seed nitrate treatments, we compared their profile in our experiments to the one they displayed in experiments comparing the transcriptome of imbibed D and ND seeds. The latter experiments analyzed the effect of nitrate or cold treatment on D seeds in the Arabidopsis Cape Verde Islands accession (Cvi; Cadman et al., 2006; Finch-Savage et al., 2007) or the effect of after-ripening or stratification in Landsberg erecta (Ler; Yamauchi et al., 2004; Carrera et al., 2007). In addition, we also included an experiment analyzing genes regulated by 30 min of nitrate provision to nitrogen-starved plants (Scheible et al., 2004).
High-nitrate feeding of mother plants led to the production of seeds with a profile of ND or AR seeds. Indeed, for C50w versus C10w, the pattern of expression of the 92 differentially expressed genes that were also present on the Affymetrix chips was correlated (P < 0.01 using Spearman's correlation test) with the pattern of these genes in the experiments studying dormancy-releasing treatments (stratification, imbibition in nitrate, after-ripening; Cadman et al., 2006; Finch-Savage et al., 2007) and shared little similarity with the transcriptomes analyzing nitrate provision (Fig. 2, top; Supplemental Table S1). Thus, most of the genes down-regulated in C50w seeds versus C10w seeds corresponded to genes higher expressed in D Ler seeds than in AR Ler seeds, whereas most of the up-regulated genes in C50 seeds were also up-regulated in AR Ler seeds.
In the nia10w versus C10w experiment, the profile of the differentially expressed genes matched that of Ler AR seeds (P < 0.05) as what had been observed in the C50w versus C10w experiment (Fig. 2, middle; Supplemental Table S2). Interestingly, there was a high representation of stress-related genes (encoding heat shock proteins) that were all but one down-regulated in the nia10w seeds compared with C10w seeds (Supplemental Table S2).
Exogenous nitrate led to the overexpression of 27 genes and underexpression of one gene in nitrate-imbibed compared with water-imbibed C10 seeds (Fig. 2, bottom; Supplemental Table S3). Many overexpressed genes were involved in nitrogen and carbon metabolism as well as energy production. The pattern of the differentially expressed genes (27 genes out of the 28 were present on the Affymetrix chip) matched significantly (P < 0.05) the profile found in experiments involving 30 min of nitrate supplementation to nitrogen-starved plants (Fig. 2) but not significantly that of the transcriptomes linked with dormancy-releasing treatments, in contrast to what had been observed for the experiments studying the effect of endogenous nitrate.
Thus exogenous nitrate led to a distinct profile from that of endogenous nitrate. In addition, although both the nia1 nia2 mutant and high-nitrate feeding of mother plants yielded nitrate-rich seeds with a ND profile, the subsets of genes differentially expressed in both types of seeds were quite distinct.
Imbibition of Wild-Type Seeds in Nitrate Increased CYP707A2 Gene Expression
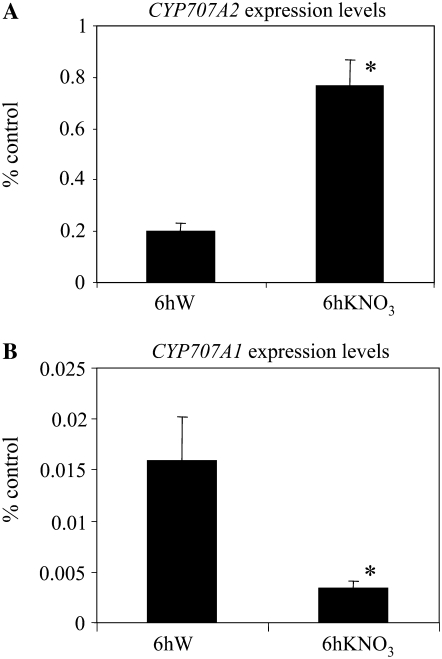
Since nitrate treatment was correlated with lower ABA levels in seeds, microarray data were inspected for the differential expression of genes involved in ABA synthesis or degradation in nitrate-treated seeds. Only the ABA biosynthesis SDR1 (ABA2) and the ABA catabolism CYP707A2 mRNAs were detected with the CATMA arrays in our samples. Of these two genes, only the CYP707A2 gene involved in ABA catabolism was overexpressed in C10 seeds imbibed for 6 h in nitrate compared with the same seeds imbibed for 6 h in water.
qRT-PCR was carried out on RNAs isolated from C10 seeds imbibed for 6 h in water or in 10 mm nitrate. CYP707A2 expression was induced about 5-fold in 6-h-imbibed C10 seeds in the presence of 10 mm exogenous nitrate compared with imbibition in water (Fig. 3A), and this induction was correlated with lower ABA levels at this time point in nitrate-imbibed seeds as assessed by liquid chromatography-tandem mass spectrometry (LC-MS/MS; see below). In addition, the CYP707A2 gene was induced also at the vegetative stage by nitrate provision to nitrogen-starved or ammonium-grown plants (Supplemental Table S3; Scheible et al., 2004), highlighting the fact that it was a nitrate-responsive gene. Therefore, we speculated that the CYP707A2 gene could be a good candidate gene that upon exogenous nitrate treatment of seeds would be induced and lead to lower dormancy of seeds due to higher ABA degradation. Both the CYP707A1 and CYP707A2 genes were reported to be important in controlling seed dormancy (Okamoto et al., 2006). The CYP707A1 gene was not detected in our microarray experiments due to its low levels of expression in mature and 6-h-imbibed seeds (Kushiro et al., 2004). To assess whether nitrate controlled the expression of the CYP707A1 gene, we analyzed its expression by qRT-PCR, a more sensitive method. CYP707A1 gene expression was indeed low in imbibed seeds and, in contrast to the CYP707A2 gene, it was repressed upon imbibition of seeds in nitrate (Fig. 3B) and was not up-regulated by nitrate provision to starved plants (Scheible et al., 2004).
Figure 3.
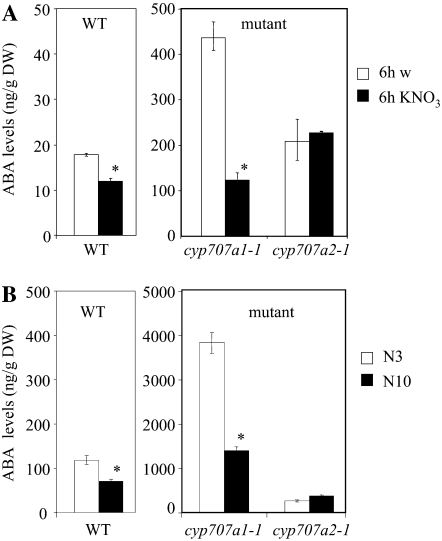
CYP707A1 and CYP707A2 mRNA levels in C10 seeds imbibed in water or nitrate. CYP707A2 (A) and CYP707A1 (B) mRNA levels were determined in C10 seeds imbibed for 6 h in water (6hW) or in 10 mm KNO3 (6hKNO3) by qRT-PCR and normalized with the expression level of the EF1α standard. Error bars indicate se (n = 3–6). For both genes, expression levels were significantly different in nitrate-imbibed (*) versus water-imbibed seeds (P < 0.05) as determined by Student's t test.
The cyp707a2 But Not the cyp707a1 Mutants Displayed Altered Dormancy Response to Exogenous Nitrate
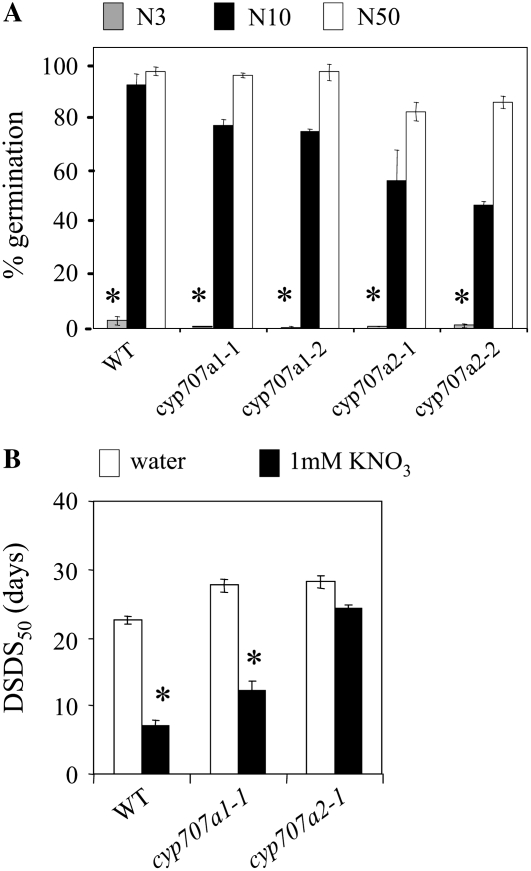
To test the role of the CYP707A1 and CYP707A2 genes in seed dormancy response to nitrate, we analyzed the response to nitrate of seeds from mutants (cyp707a1-1, cyp707a1-2, cyp707a2-1, and cyp707a2-2) harboring a T-DNA insertion in the CYP707A1 or CYP707A2 genes. In a first experiment, mother plants were cultivated in a growth chamber with 3, 10, or 50 mm nitrate nutrition (leading, respectively, to the production of N3, N10, or N50 seeds). For all genotypes, dormancy was higher (P < 0.05) in N3 seeds than in N10 seeds as assessed by germination percentage in water 10 d after harvest (Fig. 4A). Thus, wild-type and mutant seeds all responded to endogenous nitrate.
Figure 4.
Dormancy assessment of wild-type (WT), cyp707a1, and cyp707a2 mutant seeds. A, Percentage of germination in water of wild-type, cyp707a1-1, cyp707a1-2, cyp707a2-1, cyp707a2-2 N3 (gray bars), N10 (black bars), or N50 (white bars) seeds 10 d after harvest. se is indicated (n = 4). For all genotypes, there was a significant difference (P < 0.05) between the germination percentage of N3 (*) and N10 seeds as determined by ANOVA followed by Fisher's lsd test. B, DSDS50 of wild-type, cyp707a1-1, and cyp707a2-1 N10 seeds assessed in water (white bars) or in 1 mm nitrate (black bars). se is indicated (n = 6). For all genotypes except cyp707a2-1, DSDS50NO3 was significantly lower (*) than DSDS50w (P < 0.05), as determined from statistical analysis using the Wilcoxon test on paired samples.
To analyze the response to exogenous nitrate, we performed a second culture in which mother plants were cultivated with 10 mm nitrate nutrition and sowed seeds in water or in 1 mm nitrate instead of 10 mm nitrate. We reasoned that a lower concentration of nitrate would help us detect more readily small differences in responses to exogenous nitrate, as shown previously when analyzing the response to exogenous nitrate of mutants affected in the nitrate transporter genes NRT1.1 and NRT2.1 (Alboresi et al., 2005). In addition, to better assess seed dormancy, instead of analyzing seed germination right at or 1 to 2 weeks after harvest, we determined the number of days of seed dry storage necessary to obtain 50% germination (DSDS50; Alonso-Blanco et al., 2003) in water (DSDS50w) or 1 mm nitrate (DSDS50NO3). Indeed, during seed storage, dormancy is lost and the longer it takes for seeds to lose dormancy, the more D they are (the higher the DSDS50). This parameter is valuable, for example, to distinguish D seeds from very D seeds, although both types of seeds show very low percentages of germination at harvest. In addition, seeds were stored dry at 4°C to slow the after-ripening process to allow a better determination of the DSDS50 in seeds coming from N10 plants. For both wild-type and cyp707a1-1 mutant seeds, DSDS50NO3 was significantly lower than DSDS50w (P < 0.05; Fig. 4B). This confirmed their responsiveness to exogenous nitrate. For the cyp707a2-1 mutant, however, there was no significant difference (P = 0.26) between the two parameters (Fig. 4B). Thus, the cyp707a2-1 mutant but not the cyp707a1-1 mutant was affected in its response to exogenous nitrate.
ABA Levels Did Not Respond to Exogenous or Endogenous Nitrate in the cyp707a2-1 Mutant
Since ABA levels in wild-type seeds were decreased by both exogenous and endogenous nitrate treatments (Fig. 1), we analyzed ABA levels by LC-MS/MS in nitrate-treated and nontreated cyp707a1-1 and cyp707a2-1 mutant seeds. For both cyp707a1-1 and cyp707a2-1 mutants, ABA levels were higher than those in wild-type seeds either in dry or imbibed seeds as expected, since these mutants were impaired in ABA catabolism (Okamoto et al., 2006). Interestingly, imbibition of N10 seeds for 6 h in nitrate led to a significant decrease in ABA levels in both wild-type and cyp707a1-1 seeds but not for the cyp707a2-1 mutant seeds (Fig. 5A). In dry seeds, ABA levels were significantly higher in N3 than in N10 seeds for the wild type (Fig. 5B), extending our data presented in Figure 1 to N3 seeds and thus confirming that endogenous nitrate indeed affected wild-type seed ABA levels. This increase in ABA levels in N3 versus N10 seeds was also observed in the cyp707a1-1 mutant but not for the cyp707a2-1 mutant (Fig. 5B), although seed nitrate content did not differ between wild-type seeds and the two mutant seeds (data not shown). Thus, the cyp707a2-1 but not the cyp707a1-1 mutant was affected in the response of seed ABA levels to exogenous and endogenous nitrate.
Figure 5.
ABA levels in wild-type (WT) and mutant seeds treated with nitrate. A, ABA levels in N10 seeds imbibed 6 h in water (w) or in 10 mm KNO3. B, ABA levels in dry N3 and N10 freshly harvested seeds from the wild type, cyp707a1-1, and cyp707a2-1 mutants. Error bars indicate se (n = 3–4). The ABA levels for all genotypes except the cyp707a2-1 mutant were significantly lower (*) in nitrate-treated seeds than in nontreated seeds (P < 0.05), as determined by the Wilcoxon test on paired samples (A) or the Mann-Whitney test (B). DW, Dry weight.
The CYP707A2 Gene Responded to Mother Plant Nutrition during Seed Development
Although the seed dormancy of the cyp707a2-1 mutant responded to the mother plant nitrate regime (Fig. 4A), ABA levels in the mutant seeds did not respond to endogenous nitrate (Fig. 5B). Therefore, we wondered whether the CYP707A2 gene could be involved in the control of mature seed ABA levels by mother plant nitrate nutrition. Transcriptome analyses showed that in dry seeds no changes in CYP707A2 gene expression could be detected between C10 and C50 seeds. This was also confirmed by qRT-PCR in C3 and C10 dry seeds (data not shown), which displayed similar levels of CYP707A2 mRNAs.
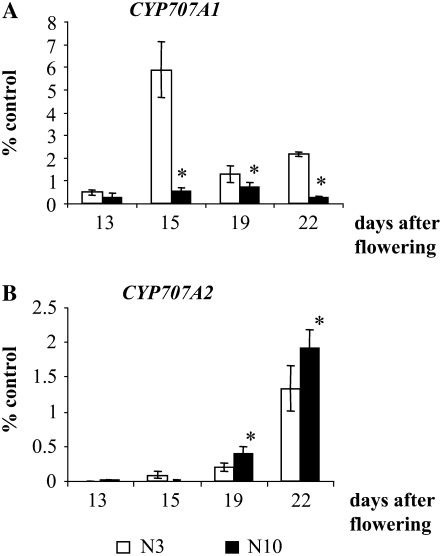
Seed primary dormancy sets up during seed maturation on mother plants. Previous studies had shown that the CYP707A1 and CYP707A2 genes were expressed during seed development at mid maturation phase for the former gene and at the late maturation phase for the latter gene (Okamoto et al., 2006). Since ABA levels in dry mature seeds depended on mother plant nutrition (Figs. 1A and 5B), we analyzed how the nitrate regime of mother plants affected CYP707A1 and CYP707A2 gene expression in developing siliques at the later stages of seed maturation. CYP707A1 mRNA levels decreased at the later stages of seed maturation and were higher in N3 than in N10 plants (Fig. 6A). In contrast, CYP707A2 expression increased at 19 and 22 d after flowering and was higher in N10 than in N3 plants (Fig. 6B). Thus, the lower levels of ABA found in mature N10 seeds versus N3 seeds were correlated with a higher expression of the CYP707A2 gene during the late maturation stage of siliques. Thus, although cyp707a2-1 seed dormancy responded to endogenous nitrate, our data indicated a possible role for CYP707A2 in controlling seed ABA levels in response to endogenous nitrate.
Figure 6.
CYP707A1 and CYP707A2 gene expression levels in developing siliques from N3 and N10 plants. A, CYP707A1 expression in developing siliques from N3 and N10 plants. B, CYP707A2 gene expression in developing siliques from N3 and N10 plants. CYP707A1 and CYP707A2 mRNA levels were determined by qRT-PCR and expressed as percentages of the control EF1α gene. Error bars indicate se (n = 3). Siliques (13–22 d after flowering) were harvested from N3 (white bars) and N10 (black bars) plants. Expression levels for both genes were significantly different (*, P < 0.05) in N3 and N10 siliques (at 15–22 d after flowering for CYP707A1 and at 19 and 22 d after flowering for CYP707A2), as determined by ANOVA followed by Fisher's lsd test.
DISCUSSION
Nitrate Treatment of Seeds Was Correlated with Lower Levels of ABA in Seeds
In this study, we analyzed the effect of exogenous and endogenous nitrate on seed dormancy by assessing ABA levels in nitrate-treated and nontreated seeds. Both exogenous and endogenous nitrate treatments led to lower ABA levels in seeds. Thus, altogether, our data point out that there is a good correlation between the lower dormancy resulting from nitrate treatments and seed ABA levels, which is consistent with results from previous studies analyzing the effect of exogenous nitrate in Cvi seeds (Ali-Rachedi et al., 2004).
Other studies reported that exogenous nitrate led likely to changes in the synthesis of GAs (Hilhorst and Karssen, 1988). Our transcriptome analyses on seeds imbibed in nitrate failed to uncover a significant change in GA biosynthesis or inactivation genes, possibly because many GA-related genes were too lowly expressed to be detected at our early time point of imbibition. In 6-h-imbibed nia10 seeds, the biosynthetic GA20ox3 gene was overexpressed (Supplemental Table S2) compared with imbibed C10 seeds. Thus, for nia10 seeds, some part of the effect of endogenous nitrate may be linked to changes in GA synthesis. More generally, it is possible that the lower ABA levels in nitrate-treated seeds combined with the cross talk between GA and ABA metabolism could lead to altered levels of GAs in nitrate-treated seeds, as shown in the light and high-temperature regulations of seed ABA levels (Seo et al., 2006; Toh et al., 2008).
Transcriptome Analyses Revealed Distinct Profiles for Nitrate-Treated Seeds
Transcriptome analyses showed that exogenous nitrate led to a distinct profile from the profiles generated by endogenous nitrate: differentially expressed genes were essentially nitrate-regulated genes involved in metabolism. It may seem surprising that in our studies the profile of C10NO3 versus C10w seeds was more similar to that of nitrogen-starved plants provided with nitrate than that of Cvi seeds imbibed in the light in the presence of nitrate. Possibly, differences in imbibition time (6 h for our analyses versus 24 h for Cvi seeds) or in the genetic background (Col-0 versus Cvi) or in the degree of dormancy could explain these discrepancies. Endogenous nitrate resulted in a profile at 6 h of imbibition similar to that of ND seeds (cold treated or AR or imbibed in the presence of nitrate), but the profile associated with C50w versus C10w was distinct from that obtained in nia10w versus C10w, with few common differentially expressed genes. Thus, with respect to transcriptome analyses, the ND C10NO3, C50w, and nia10w seeds seem to represent quite distinct physiological states. The differences in the mRNA profiles of nitrate-treated seeds may arise from the experimental design of the nitrate treatments, since the effect of endogenous nitrate is exerted throughout seed development, maturation, and imbibition, in contrast to the effect of exogenous nitrate, which operates only during the 6 h of seed imbibition. Likewise, although the nia10 seeds accumulate nitrate like C50 seeds (Alboresi et al., 2005), the nia1 nia2 mutant is partially impaired in nitrate assimilation, which may explain its distinct transcriptome profile. The numerous heat shock protein-encoding genes that were down-regulated in the nia10 seeds were almost all down-regulated during a germination time course at 24 h of imbibition (Nakabayashi et al., 2005). Thus, the ND nia10 seeds appear to have accelerated the decrease in heat shock gene expression during imbibition.
A Central Role for the CYP707A2 Gene in Nitrate Control of ABA-Regulated Seed Dormancy
Our work furthermore stresses the central role of the CYP707A2 gene in controlling seed dormancy in response to nitrate. Indeed, transcriptome analyses and qRT-PCR experiments showed that exogenous nitrate led to an increase in the mRNA levels of the ABA catabolic gene CYP707A2 in imbibed seeds. Physiological analyses showed that a mutation in the CYP707A2 gene alone was sufficient to abolish or greatly reduce seed response to exogenous nitrate. This indicated that the CYP707A2 gene played a major role in mediating control by exogenous nitrate of seed ABA levels and dormancy. Thus, our work highlighted the importance of the regulation of ABA degradation more than ABA synthesis in controlling seed dormancy response to exogenous nitrate. Work by Bethke et al. (2006a) showed that nitrate, nitrite, and cyanide broke seed dormancy via the production of NO and changes in ABA sensitivity but not ABA synthesis. Our data here could explain the changes in ABA sensitivity upon imbibition in nitrate, since the induction of an ABA-degrading enzyme by nitrate would lower a seed's sensitivity to exogenous ABA. Indeed, we did observe that seeds imbibed in nitrate were slightly more resistant to exogenous ABA than water-imbibed seeds (data not shown).
Our data furthermore highlighted the possible involvement of the CYP707A2 gene in the control by endogenous nitrate of mature seed ABA content. Indeed, the expression profile of the CYP707A2 gene was consistent with its involvement in determining mature seed ABA levels in response to mother plant nitrate nutrition. Mature seed ABA levels in the cyp707a2-1 mutant remained unchanged when mother plant nutrition was altered, in contrast to wild-type seeds. Dormancy of the mutant, however, responded to endogenous nitrate and was higher for lower mother plant nitrate nutrition. This indicated that the control by endogenous nitrate of seed dormancy also involved other processes than just the regulation of dry seed ABA levels. The importance of regulating seed ABA levels during imbibition for seed dormancy was stressed by our studies as well as previous ones (Ali-Rachedi et al., 2004). It is possible that other genes than the CYP707A2 gene are involved in the control by endogenous nitrate of seed dormancy. Indeed, the ABA synthesis NCED9 gene appears to be higher expressed in C3 than in C10 seeds during seed imbibition in water (T. Matakiadis, unpublished data). The dual control of ABA levels by an ABA synthesis gene and an ABA catabolism gene during seed imbibition could explain why the dormancy of the cyp707a2-1 mutant still responded to endogenous nitrate.
Our work underlines also the nonredundant roles of the CYP707A1 and CYP707A2 genes. Indeed, the cyp707a1-1 mutant remained responsive to exogenous nitrate, in contrast to the cyp707a2-1 mutant, stressing the specific roles of the different members of the small multigenic family encoding ABA 8′-hydroxylases in Arabidopsis (Okamoto et al., 2006; Umezawa et al., 2006). The CYP707A2 gene was proposed to be important for a seed's response to light (Seo et al., 2006). This, altogether with our data, stresses the important role of the CYP707A2 gene in regulating seed ABA levels and dormancy in response to environmental cues such as nitrate, light, and storage time. Comparison of our transcriptome data with published data on nitrate provision to nitrogen-starved seedlings (Scheible et al., 2004) highlighted the fact that the CYP707A2 gene was nitrate inducible even in processes not linked with dormancy. Data from Wang et al. (2004) studying nitrate-regulated genes uncovered a set of about 600 genes that responded to nitrate in both the wild type and a nitrate reductase null mutant. This set of genes, which included the CYP707A2 gene, was classified as responsive to nitrate per se, since the response to nitrate was independent of nitrate reductase activity. The CYP707A2 gene can thus be classified as a bona fide nitrate-inducible gene. The CYP707A1 gene, in contrast, was not induced by exogenous nitrate in seeds: on the contrary, it was repressed upon imbibition of seeds in nitrate and repressed in developing siliques by higher mother plant nutrition. Thus, its contribution to modulating ABA levels in these conditions would be limited, in contrast to that of the CYP707A2 gene. Summarizing our data, a simplified model would thus be that nitrate (provided either exogenously or endogenously) would accumulate in seeds through the action of nitrate transporters (possibly NRT1.1 for nitrate transport into seed/embryo; Alboresi et al., 2005), then accumulate in the storage vacuoles by the action of the nitrate transporter NRT2.7 (Chopin et al., 2007). Nitrate then would induce the expression of the CYP707A2 gene, leading to changes in ABA levels through ABA degradation and lowered ABA synthesis and thus lower seed dormancy. Interestingly, the nitrate inducibility of the CYP707A2 gene in leaves and that of the CYP707A3 gene in roots (Wang et al., 2004) raises the question of what links exist between nitrate and ABA during plant development at the vegetative stage. Nitrate and ABA were already reported to interact in regulating root architecture (Signora et al., 2001; De Smet et al., 2003). It is possible that more generally these two pathways interact to adapt plants to nutritional cues.
Although our data point to a major role of the CYP707A2 gene in mediating nitrate control of seed dormancy, not all of the effect of nitrate was mediated through this gene. Indeed, in some experiments, the cyp707a2 mutant seeds still responded partially to nitrate (1 mm) in the medium and responded normally to 10 mm exogenous nitrate. The partial phenotype of this mutant could be due to overlapping roles of CYP707A2 and other genes such as NCED9, which could also be involved in dormancy control by nitrate. Alternatively, nitrate could affect seed dormancy through changes other than transcriptional changes. The importance of posttranscriptional and translational control of seed germination was indeed underlined in other studies (Rajjou et al., 2004; Chibani et al., 2006). In addition, metabolic changes in seed composition or during seed imbibition after nitrate treatment could contribute to these changes in seed germination. Our transcriptome analyses indicated that exogenous nitrate did indeed lead to an increase in the expression of genes involved in nitrogen and carbon metabolism as well as in energy production. Since ABA was reported to inhibit seed germination by restricting the availability of metabolites and energy (Garciarrubio et al., 1997), it is possible that nitrate treatment led to metabolic changes enabling the seed to overcome this inhibition. Metabolomic analyses of nitrate-treated seeds are under way to better understand the impact of mother plant nutrition and seed metabolism during imbibition on seed composition and germinative quality.
MATERIALS AND METHODS
Plant Material and Seed Production
Arabidopsis plants (Arabidopsis thaliana Col-0 and the double mutant G'4-3 [nia1nia2]; Wilkinson and Crawford, 1993) in the Col-0 background were used in this study, as well as the cyp707a1-1, cyp707a1-2, cyp707a2-1, cyp707a2-2, cyp707a3-1, and cyp707a3-2 (Kushiro et al., 2004, Okamoto et al., 2006) mutants. For seeds produced under controlled conditions, seeds were stratified for 48 h at 4°C in the dark in a 1% agar solution. Approximately five seeds were sown in a small pot filled with homogenous nonenriched compost. One week after sowing, only one seedling per pot was retained. Plants were grown in a growth chamber at 22°C/18°C under a 16-h photoperiod of artificial light (100 μE m−2 s−1) and 80% relative humidity. The pots were watered three times per week by immersion of the base of the pots in a solution containing either 3 or 10 mm nitrate. One week after bolting, the nitrate regime of a few plants watered with 10 mm nitrate was changed to 50 mm nitrate, as described initially by Alboresi et al. (2005).
Siliques of different developmental stages were obtained by tagging newly opened flowers every 3 d.
Seed Dormancy and Germination Assays
For each experiment, all genotypes grown with the various nitrate regimes (3, 10, and 50 mm nitrate) were harvested when siliques turned dry on the plants. For dormancy analysis, freshly harvested seeds from at least four independent seed lots (each obtained by pooling seeds produced by three to four mother plants) were sown on 0.5% agarose plates (Litex agarose; Vallensbaek Strand) with or without 1 mm KNO3. The plates were incubated in a growth chamber (Cu-36L6; Percival) at 25°C with 16 h of light (100 μE m−2s−1) and 8 h of dark.
For DSDS50 determinations (Alonso-Blanco et al., 2003), seeds were stored dry at 4°C in screw-cap tubes and sown to determine the percentage of germinating seeds of each genotype after different storage times. DSDS50 (Alonso-Blanco et al., 2003) was then determined when possible by probit regression on a logarithm time scale applying the XLSTAT dose module of the XLSTAT software (Addinsoft France).
qRT-PCR
Total RNA was isolated using an RNAqueous column with the Plant RNA isolation aid (Ambion) followed by a two-step LiCl precipitation. The isolated RNA was mixed with one-quarter volume of 10 m LiCl and stored at −80°C overnight. The sample was then allowed to warm to room temperature and centrifuged at 5,000g at 4°C for 15 min. The pellet was washed with 50 μL of 2 m LiCl, incubated on ice for 10 min, and centrifuged at 5,000g at 4°C for 10 min. A final wash step was performed with 70% (v/v) ethanol, and then samples were centrifuged at 5,000g at 4°C for 5 min. After removal of the supernatant, the pellet was resuspended in 12 μL of diethyl pyrocarbonate-treated double-distilled water and stored at −80°C.
First-strand cDNA was synthesized with random hexamers using a SuperScript first-strand synthesis system as described by Daniel-Vedele and Caboche (1993) using Moloney murine leukemia virus reverse transcriptase and random primers (Promega) or using the SuperScript first-strand synthesis system according to the manufacturer's instruction (Invitrogen). qRT-PCR was carried out using the first-strand cDNA as a template using the Mastercycle instrument (Mastercycle ep realplex; Eppendorf) with the RealMasterMix kit (Eppendorf) according to the manufacturer's protocol. Primers used for qRT-PCR for the CYP707A1, CYP707A2, and EF1α genes were those described previously (Orsel et al., 2004; Seo et al., 2004; Millar et al., 2006).
Determination of ABA Levels
Preliminary measurements of seed ABA contents (Fig. 1) were done by HPLC followed by ELISA as described by Lefebvre et al. (2006). For the other series of ABA determinations, seed samples were extracted as described previously (Priest et al., 2006). Purified ABA was quantified by LC-MS/MS (Q-Tof premier; Micromass) using the following mass-to-charge ratio peaks, 159 for labeled ABA and 153 for endogenous ABA. The amount of each compound was generated by spectrometer software (MassLynx version 4.1; Micromass).
Transcriptome Studies
Microarray analysis was carried out at the Unité de Recherche en Génomique Végétale using the CATMA array (Crowe et al., 2003; Hilson et al., 2004), containing 24,576 gene-specific tags from Arabidopsis. RNA samples from two independent biological replicates were used. For each biological repetition, RNA samples for a condition were obtained by extracting RNAs from seeds of four to eight plants and pooling them. For each comparison, one technical replication with fluorochrome reversal was performed for each biological replicate (i.e. four hybridizations per comparison). The RT of RNA in the presence of Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization of labeled samples to the slides, and the scanning of the slides were performed as described (Lurin et al., 2004).
Data from published experiments using Affymetrix arrays were obtained as supplemental data of published papers (Scheible et al., 2004; Carrera et al., 2007) or downloaded through The Arabidopsis Information Resource Web site (Yamauchi et al., 2004) or through the NASC Affywatch service (Finch-Savage et al., 2007) and focusing on the PDLN versus PDL and PDC versus PD24h comparisons. For data from Carrera et al. (2007; Supplemental Table I), differentially expressed genes between AR and D Ler seeds were obtained by adding to the list of up-regulated genes in Ler AR seeds (LerARupvsLerD) the list of the genes up-regulated in Ler D seeds (LerDupvsLerAR), which represent the down-regulated genes in AR Ler seeds versus D Ler seeds. For the other published microarray data comparing a test sample versus a control sample, genes were considered induced if their call values were P in all replicates of the test samples and their expression displayed at least a 2-fold change with respect to control. Genes were considered repressed compared with controls if their call value was P in all replicates of control samples and if they were expressed at least 2-fold less in test samples compared with control samples.
Statistical Analysis of Data
Experiments based on the CATMA arrays were designed with the statistics group of the Unité de Recherche en Génomique Végétale. Statistical analysis was based on two dye swaps (i.e. four arrays, each containing 24,576 gene sequence tags and 384 controls) as described (Gagnot et al., 2008). Controls were used for assessing the quality of the hybridizations but were not included in the statistical tests or the graphic representation of the results. Data normalization and determination of differentially expressed genes by paired t test on the log ratios using a Bonferroni P value of <5% were as described by Merigout et al. (2008).
The profile of genes differentially expressed in nitrate-treated seeds versus nontreated seeds was compared with the profile in published experiments using Affymetrix chips by performing a Spearman correlation exact test with the Monte Carlo approximation (10,000 samples) and Bonferroni's correction in the case of multiple comparisons.
For statistical analysis of ABA levels, germination percentages, DSDS50, and qRT-PCR data parametric tests (Student's t test, ANOVA followed by Fisher's lsd test) were performed when the different samples displayed equal variances (which was in some cases obtained after log transformation of data). Otherwise, nonparametric tests (Mann-Whitney or Wilcoxon tests) were used to test the significance of the results.
Microarray data from this article were deposited in the Array Express (http://www.ebi.ac.uk/arrayexpress/; accession no. E-MEXP-447) and the CATdb (http://urgv.evry.inra.fr/CATdb/; Project RA04-05_Dormancy-NO3) databases according to Minimum Information about a Microarray Experiment standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genes differentially expressed in nitrate-treated imbibed seeds.
Supplemental Figure S2. qRT-PCR analysis of a set of genes differentially expressed in nitrate-treated imbibed seeds.
Supplemental Table S1. Genes differentially expressed in C50w versus C10w seeds and their profiles in nitrate and seed transcriptomes.
Supplemental Table S2. Genes differentially expressed in nia10w versus C10w seeds and their profiles in nitrate and seed transcriptomes.
Supplemental Table S3. Genes differentially expressed in C10NO3 versus C10w seeds and their profiles in nitrate and seed transcriptomes.
Supplemental Table S4. Genes differentially expressed in C50ds versus C10ds seeds.
Supplemental Table S5. Genes differentially expressed in nia10ds versus C10ds seeds.
Supplemental Table S6. Differentially expressed genes common to C50ds versus C10ds and niads versus C10ds experiments.
Supplemental Table S7. Differentially expressed genes common to C50w versus C10w and C50ds versus C10ds experiments.
Supplemental Table S8. Differentially expressed genes common to niaw versus C10w and niads versus C10ds experiments.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of B. Sotta and S. Lachaud for determining ABA levels in seeds by HPLC followed by immunodetection, Y. Kanno for extraction and purification of ABA for analysis by LC-MS/MS, M. Bedu and A. Weinbach for help with seed harvest and sampling, and J. Talbotec and F. Gosse for care of plants in the greenhouse and in growth chambers. We thank M. Okamoto for discussion and comments on the experiments and A. Marion-Poll, C. Meyer, and F. Daniel-Vedele for critical reading of the manuscript and helpful discussions.
This work was supported by the European Union Early Stage Training Site VERT (grant no. MEST–CT–2004–7576 VERT to T.M.) and by the European Union Fifth Framework Research Training Network PLUSN (grant no. UE HPRN CT 00247 to A.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hoai-Nam Truong (truong@versailles.inra.fr).
The online version of this article contains Web-only data.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28 500–512 [DOI] [PubMed] [Google Scholar]
- Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219 479–488 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batak I, Devi M, Giba Z, Grubisi D, Poff KL, Konjevic R (2002) The effects of potassium nitrate and NO-donors on phytochrome A- and phytochrome B-specific induced germination of Arabidopsis thaliana seeds. Seed Sci Res 12 253–259 [Google Scholar]
- Bentsink L, Koornneef M (2002) Seed dormancy and germination. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–18
- Bethke PC, Libourel IGL, Jones RL (2006. a) Nitric oxide reduces seed dormancy in Arabidopsis. J Exp Bot 57 517–526 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Reinohl V, Jones RL (2006. b) Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223 805–812 [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46 805–822 [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ (2008) Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J 53 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ (2007) Gene expression profiling reveals defined functions of the ATP-binding cassette transporter comatose late in phase II of germination. Plant Physiol 143 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P (2006) Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol 142 1493–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel-Vedele F (2007) The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Vedele F, Caboche M (1993) A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol Gen Genet 240 365–373 [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inze D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33 543–555 [DOI] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J (2005) The evolutionary ecology of seed germination of Arabidopsis thaliana: variable natural selection on germination timing. Evolution Int J Org Evolution 59 758–770 [PubMed] [Google Scholar]
- Donohue K, Heschel MS, Butler CM, Barua D, Sharrock RA, Whitelam GC, Chiang GCK (2008) Diversification of phytochrome contributions to germination as a function of seed-maturation environment. New Phytol 177 367–379 [DOI] [PubMed] [Google Scholar]
- Donohue K, Heschel MS, Chiang GCK, Butler CM, Barua D (2007) Phytochrome mediates germination responses to multiple seasonal cues. Plant Cell Environ 30 202–212 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51 60–78 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36 D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203 182–187 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8 183–187 [DOI] [PubMed] [Google Scholar]
- Hayes GR, Klein WH (1974) Spectral quality influence of light during development of Arabidopsis thaliana plants in regulating seed germination. Plant Cell Physiol 15 643–653 [Google Scholar]
- Hilhorst HWM, Karssen CM (1988) Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol 86 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM, Karssen CM (1989) Nitrate reductase independent stimulation of seed germination in Sisymbrium officinale L. (hedge mustard) by light and nitrate. Ann Bot (Lond) 63 131–137 [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45 309–319 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough JM, Shropshire WJ (1970) Physiological predetermination of germination response in Arabidopsis thaliana (L). Heynh. Plant Cell Physiol 11 139–148 [Google Scholar]
- Merigout P, Gaudon V, Quillere I, Briand X, Daniel-Vedele F (2008) Urea use efficiency of hydroponically grown maize and wheat. J Plant Nutr 31 427–443 [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45 942–954 [DOI] [PubMed] [Google Scholar]
- Munir J, Dorn LA, Donohue K, Schmitt J (2001) The effect of maternal photoperiod on seasonal dormancy in Arabidopsis thaliana (Brassicaceae). Am J Bot 88 1240–1249 [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode ABA 8′-hydroxylases, are indispensable for a proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219 714–721 [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA (2005) Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15 1998–2006 [DOI] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross ARS, Abrams SR, Bowles DJ (2006) Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J 46 492–502 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB (1997) Gibberellin mutants. Physiol Plant 100 550–560 [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45 1694–1703 [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48 354–366 [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28 655–662 [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, et al (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146 1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46 171–182 [DOI] [PubMed] [Google Scholar]
- Wang RC, Tischner R, Gutierrez RA, Hoffman M, Xing XJ, Chen MS, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1993) Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet 239 289–297 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y (2002) Gibberellins and light-stimulated seed germination. J Plant Growth Regul 20 369–376 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.