Abstract
Double-stranded (ds)RNA interference (RNAi) is widely used for functional analysis of plant genes and is achieved via generating stable transformants expressing dsRNA in planta. This study demonstrated that RNAi can also be utilized to examine gene functions in protoplasts. Because protoplasts are nongrowing cells, effective RNAi-triggered gene silencing depends not only on a depletion of gene transcripts but also on turnover rates of corresponding polypeptides. Herein, we tested if transient RNAi in protoplasts would result in the depletion of a targeted polypeptide and, because protoplasts have a limited life span, if functional assays of RNAi knockout genes would be feasible in protoplasts. We showed that protoplasts transfection with an in vitro-synthesized dsRNA against Arabidopsis (Arabidopsis thaliana) β-glutamylcysteine synthase (ECS1), a key enzyme in the synthesis of glutathione, resulted in a 95% depletion of ECS1 transcript, a 72% decrease of ECS1 polypeptide, and a 60% drop in glutathione content. These results were comparable with those obtained upon analysis of Arabidopsis seedlings bearing the cad2-1 mutant allele of ECS1. We also improved the procedure for RNAi inactivation of several genes simultaneously. Finally, because we isolated protoplasts from tissues of 14-d-old seedlings instead of 1-month-old mature plants, the described procedure is rapid (as it only takes 20 d from seed planting to functional studies), suitable for analyzing multiple genes in parallel, and independent of cloning dsRNAs into plant expression vectors. Therefore, RNAi in protoplasts complements existing genetic tools, as it allows rapid, cost- and space-efficient initial screening and selection of genes for subsequent in planta studies.
The completion of the Arabidopsis Genome Sequencing Project opened a new era in plant biology research, the challenges of which are to determine the functions of all annotated Arabidopsis (Arabidopsis thaliana) genes and to extend these studies to other plant species (AGI, 2000). Indeed, although the functions of 69% of the genes were classified according to sequence similarity to proteins of known function in all organisms, only 16,000 of the 29,000 predicted genes have been characterized experimentally, and these studies have been done primarily in Arabidopsis (AGI, 2000; Wortman et al., 2003). A critical step in functional analysis of plant genes is the availability of genetic tools that are applicable to different plant species, are rapid, affordable, and thus can be used as the initial step in the genome-wide screens and selection of genes with traits of interest for subsequent in-depth assessment in planta.
Double-stranded (ds)RNA interference (RNAi) is an RNA-based reverse-genetic approach currently in use for studies of gene function. RNAi silencing is triggered by the introduction of dsRNA into cells where it is detected as aberrant and is processed by the type III RNase Dicers to small, 20- to 26-nucleotide-long, short interfering RNAs (siRNAs; Bernstein et al., 2001; Brodersen and Voinnet, 2006). One of the two siRNA strands is then incorporated into the RNA-induced-silencing complex, which uses siRNAs to recognize complementary motifs in target nucleic acids. The result is the sequence-specific inhibition of gene expression either at the transcription, mRNA stability, or translational levels (Baulcombe, 2004; Brodersen and Voinnet, 2006; Brodersen et al., 2008). The sequence specificity of RNAi-based gene inactivation allows silencing of individual genes as well as several genes simultaneously. Therefore, RNAi can be used to silence multiple members of a multigene family and homologous gene copies in polyploids by targeting sequences that are unique or shared by these related genes (Waterhouse and Helliwell, 2003; Baulcombe, 2004).
RNAi has proven to be very efficient in interfering with gene expression in various organisms, including vertebrate and invertebrate animals, and has been used for gene function studies in plant systems such as Physcomitrella patens, Petunia hybrida, Arabidopsis, Papaver somniferum, rice (Oryza sativa), wheat (Triticum aestivum), and others (Stam et al., 1997; Fire et al., 1998; Kennerdell and Carthew, 1998; Smith et al., 2000; Waterhouse et al., 2001; Waterhouse and Helliwell, 2003; Allen et al., 2004; Baulcombe 2004; Bezanilla et al., 2005; Miki et al., 2005; Travella et al., 2006; Vidali et al., 2007). In these plant species, RNAi is achieved by transforming plants with constructs that express self-complementary (hairpin) RNA-containing sequences that are homologous to targeted genes, or with constructs expressing artificial microRNAs (Waterhouse and Helliwell, 2003; Helliwell et al., 2005; Schwab et al., 2006).
The generation of the genome-wide collection of artificial microRNA-expressing plasmids and availability of transposon/T-DNA mutant alleles of Arabidopsis provided the plant research community with an outstanding resource for functional genomics (Alonso et al., 2003; http://2010.cshl.edu/scripts/main2.pl?link=project&content=project.html). Using these tools, however, requires growth and propagation of transgenic plants and thus imposes extensive time, labor, and space requirements for maintaining multiple plant lines. Therefore, the availability of a rapid and cost-efficient reverse-genetic approach for the initial genome-wide analysis and selection of a subset of genes of interest for subsequent in planta studies would greatly assist and expedite functional genetic studies.
In this regard, in animals, the development of efficient dsRNA delivery approaches has enabled studies of gene function in cell tissue cultures (Fire et al., 1998; Worby et al., 2001; Wilson and Richardson, 2003). Because of the simplicity of RNAi in animal cell cultures, minimal time and space are required for gene inactivation and subsequent phenotype analysis. In fact, this approach has been recently adapted to a genome-wide high-throughput format, is used as an initial screening step for genes of interest, and has resulted in the identification of novel genes with roles in cell growth and development, life span regulation, muscle differentiation, signal transduction pathways, and other processes (Clemens et al., 2000; Kamath et al., 2003; Boutros et al., 2004; Hamilton et al., 2005; Bai et al., 2008).
Aiming to develop RNAi for rapid functional genetic screens by circumventing the time- and space-intensive production of transgenic RNAi plants, we sought to test if, as in animal cells, RNAi could be used to study gene functions in isolated cells, i.e. protoplasts. We chose protoplasts for the following reasons: (1) protoplasts are easily prepared; (2) protoplasts are used to study a wide range of cellular functions, such as cellular transport mechanisms and the subcellular localization of proteins (Chen and Halkier, 2000); (3) protoplasts can be isolated from different parts of plants (roots, stems, leaves), thus enabling studies of tissue-specific processes (e.g. nutrient uptake in root cells and storage in leaf cells); and (4) importantly, protoplasts can be readily transfected with nucleic acids (dsRNA or DNA; An et al., 2003, 2005; Yoo et al., 2007). Lastly, it has been shown that transfection of dsRNA into Arabidopsis protoplasts reduces the transcript level of the targeted exogenous and endogenous genes (An et al., 2003, 2005). Whether reduction of the endogenous gene-transcript leads to a depletion of the corresponding polypeptide and interferes with its function has not been determined (An et al., 2003, 2005). However, this is important to establish, because protoplasts are nongrowing cells, and the interference with gene function by transient RNAi depends not only on the depletion of transcripts but also on turnover rates of corresponding polypeptides and, thus, on the RNAi depletion of targeted polypeptides. In addition, protoplasts have limited viability; therefore, it is important to determine whether the time frame of the viability of RNAi protoplasts in the artificial media is sufficient for functional analysis. These questions need to be addressed prior to developing a high-throughput RNAi protocol in protoplasts useful for functional genetics.
Herein, we showed that effective RNAi-mediated gene silencing that was achieved in Arabidopsis protoplasts was applicable for analysis of gene function. We showed that the transfection of an in vitro-synthesized dsRNA against Arabidopsis β-glutamylcysteine synthase (ECS1), a key enzyme in the synthesis of glutathione (GSH) into protoplasts resulted in the depletion of ECS1 transcript, the decline of ECS1 polypeptide, and the decline of the protoplast content of the end product of ECS1 enzymatic activity, GSH. Importantly, the fold-decrease of GSH content in RNAi protoplasts was comparable with that of the cad2-1 mutant allele of ECS1 (Cobbett et al., 1998). We also significantly improved the procedure for the efficient silencing of genes individually and homologous members of gene families simultaneously. The procedure reported in this study is rapid; only 20 d are required for the analysis of gene function. This time period includes growth of Arabidopsis, in vitro dsRNA synthesis, protoplast isolation, transfection, and analysis. Therefore, RNAi in protoplasts described here illustrated a rapid, affordable, and potent reverse-genetic approach that is complementary to existing genetic tools, as it allows expediting functional analysis of plant genes and, as detailed in “Conclusion and Future Perspectives,” is particularly applicable for biochemical studies for deciphering components of metabolic pathways, establishing of metabolic networks, and, perhaps, can be adapted to high-throughput metabolomics.
RESULTS
Isolation of Intact Protoplasts from 14-d-Old Arabidopsis Seedlings and Achieving High Protoplast Transfection Efficiency
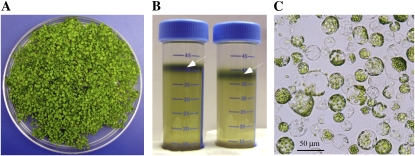
The success of RNAi silencing of genes in protoplasts is contingent on sufficient yield and viability of isolated protoplasts and their transfection efficiency. Isolation of protoplasts was first reported more than 40 years ago (Cocking, 1960) and has since been adapted to study a variety of cellular processes, such as subcellular localization of proteins and the isolation of intact organelles (Yoo et al., 2007). Most protoplast isolation protocols use leaf tissues of mature Arabidopsis (e.g. 35 d old). We modified existing protocols to decrease the time and growth chamber space that are required for isolating protoplasts from mature plants by employing 14-d-old Arabidopsis seedlings (Fig. 1A).
Figure 1.
Isolation of protoplasts from Arabidopsis seedlings. A, Tissues from 14-d-old seedlings of Arabidopsis were collected and converted to protoplasts by a modified procedure of Chen and Halkier (2000). B, Protoplasts were purified by Suc density gradient centrifugation and collected at the interface of enzyme solution and W5 buffer (white arrows). C, Bright-field microscopy of protoplasts. Microphotographs were collected using a cooled CCD camera interfaced with the Zeiss Axioscope 2 plus microscope.
Arabidopsis seedlings were collected from a petri plate, sliced with a razor blade, and converted to protoplasts in a buffer containing Macerozyme R-10 and Cellulase (see “Materials and Methods”). The released protoplasts were purified by Suc density gradient centrifugation and collected at the interface of the enzyme and W5 solutions (Fig. 1B). Using this procedure, 1 g of 14-d-old seedlings yielded 5 × 106 to 107 protoplasts (Fig. 1C). The yield of protoplasts from seedlings is comparable with preparations from leaves of mature Arabidopsis, but instead of 35 to 36 d, isolation of protoplasts was completed in 15 d. This improvement is significant, because it allowed us to decrease the time required for RNAi-based analyses in protoplasts.
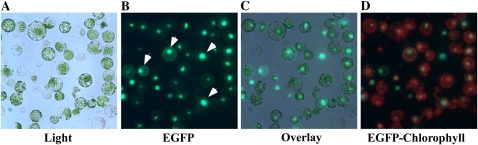
We then established an efficient transfection procedure for the protoplasts. We employed a simple visual assay to assess the transfection efficiency using fluorescence microscopy. We transfected protoplasts with the pSAT plasmid, expressing the nuclear-targeted red-shifted GFP (EGFP) driven by two cauliflower mosaic virus 35S promoters, oriented in tandem (Tzfira et al., 2005). Protoplasts were transfected using a poly(ethylene glycol) (PEG)-calcium-mediated transfection procedure as previously described (Yoo et al., 2007), except that we used 10 times more protoplasts than detailed in (Yoo et al., 2007). Fluorescent microscopy revealed that more than 90% of transfected protoplasts expressed the nuclear-targeted EGFP, suggesting that we achieved high transfection rates (Fig. 2, A–D).
Figure 2.
The visualization of the efficiency of the transfection of protoplasts with the pSAT vector expressing nuclear-localized EGFP. Bright-field (A, Light) and fluorescent microphotographs (B, EGFP) were collected on the Zeiss Axioscope 2 plus microscope equipped with appropriate filter sets. White arrows indicate examples of nuclear-localized, EGFP-mediated fluorescence in the pSAT plasmid-transfected protoplasts. C, Superimposed bright-field and fluorescent images on A and B (Overlay) were created to demonstrate the transfection efficiency. D, Overlaid images of autofluorescence from chloroplasts and fluorescence from EGFP (EGFP-Chlorophyll) was created to demonstrate that green fluorescence derives solely from EGFP of the pSAT vector.
RNAi-Mediated Silencing of the Expression of the Exogenous Gene, EGFP, in Protoplasts
We then tested if RNAi effects indeed could be observed in protoplasts and if transfecting in vitro-synthesized dsRNA, rather than a construct expressing dsRNAs in vivo, could induce RNAi effects in protoplasts (An et al., 2005).
We synthesized dsRNA against a 500-bp-long fragment corresponding to a coding sequence of EGFP (dsRNAEGFP) and cotransfected it with pSAT-EGFP into protoplasts. To evaluate the transfection efficiency, we transfected a subset of protoplasts with the pSAT-EGFP vector alone. We also transfected a subset of protoplasts with water (mock transfection) and used these protoplasts for monitoring the autofluorescence of chlorophyll.
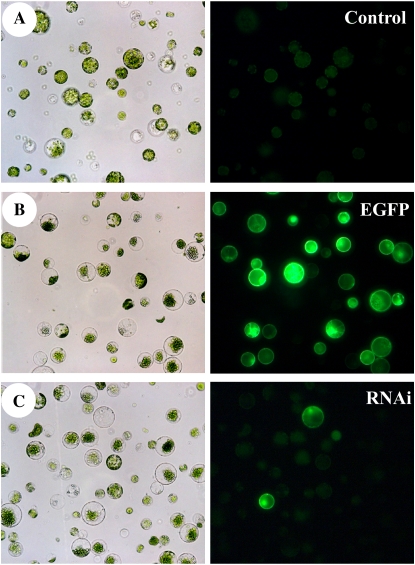
Fluorescent microscopy analysis established that mock-transfected protoplasts exhibited little or no fluorescence in the GFP-specific filter set (Fig. 3A). Weak fluorescence in mock-transfected protoplasts was attributed to chlorophyll autofluorescence. In contrast, as determined by the nuclear-localized, EGFP-mediated fluorescence, 90% of cells transfected with the pSAT-EGFP plasmid appeared to express the plasmid (Fig. 3B). The protoplasts transfection protocol, therefore, delivered nucleic acids efficiently.
Figure 3.
Silencing of the exogenous gene, EGFP, in protoplasts by RNAi. A, Bright-field (left) and fluorescence (right) micrographs of mock-transfected protoplasts (Control). B, Light (left) and fluorescence (right) micrographs of protoplasts transfected with the pSAT plasmid expressing nuclear-targeted EGFP (EGFP). C, Microphotographs of protoplasts cotransfected with the pSAT plasmid and in vitro-synthesized dsRNA against EGFP (RNAi). Fluorescence micrographs were taken after 24 h of transfection. Images were captured using the same exposure time.
We next tested if the introduction of dsRNAEGFP in conjunction with the pSAT-EGFP would affect the expression of EGFP. We determined that cotransfection with dsRNAEGFP substantially decreased the number of protoplasts that exhibited the EGFP-mediated fluorescence (Fig. 3C). Based on these data and previous observations (An et al., 2003), we concluded that indeed, transfection of an in vitro synthesized dsRNA into protoplasts triggers transient RNAi effects.
RNAi-Mediated Silencing of the Expression of the Endogenous Arabidopsis Gene, PCS1, Depends on the Dose of Transfected dsRNA
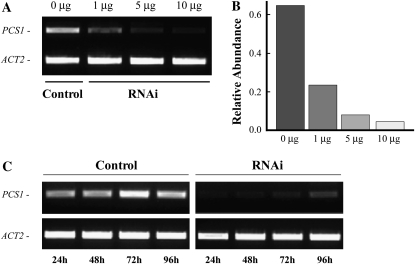
We next sought to assay the efficiency of RNAi effects on an endogenous Arabidopsis gene and to optimize the potency of RNAi effects relative to the amount of transfected dsRNA. We performed RNAi against PCS1 (for phytochelatin synthase 1), because the expression of its mRNA in Arabidopsis is well characterized (Vatamaniuk et al., 1999, 2000). The reverse transcription (RT)-PCR analysis of mock- and dsRNAPCS1-transfected protoplasts established that transfection with 1 μg of dsRNAPCS1 decreased the abundance of PCS1 transcript by 60% in comparison with the mock-transfected protoplasts (Fig. 4, A and B). The potency of RNAi effects, however, increased with a higher dose of transfected dsRNAPCS1 (Fig. 4, A and B). The abundance of PCS1 transcript in RNAi protoplasts decreased by 70% and 90% when 5 μg and 10 μg of dsRNAPCS1, respectively, were used for transfection (Fig. 4B). Because transfection of 10 μg of dsRNAPCS1 caused the most potent gene-silencing effect, we used 10 μg of dsRNAs for transfection in subsequent studies.
Figure 4.
Dose and time dependence of transient RNAi silencing of PCS1 in protoplasts. A, Protoplasts were transfected with 1, 5, or 10 μg of dsRNA against PCS1 (RNAi) or with water (Control) and incubated for 24 h in the dark. Silencing of PCS1 in dsRNA- and mock-transfected protoplasts was analyzed by RT-PCR. The representative RT-PCR results of three independent transfection events are shown. B, Relative abundance of PCS1 transcript on the gel in A. Relative abundance was normalized to the level of the transcript of ACT2 using GelQuant software. C, RT-PCR analysis of the time dependence of RNAi silencing of PCS1 in protoplasts. Protoplasts were transfected with 10 μg of dsRNA against PCS1 (RNAi) or with water (Control). Control and RNAi protoplast samples were collected at indicated time points and analyzed by RT-PCR for the abundance of PCS1 transcript. ACT2 was amplified as a control for amount of a template.
RNAi Effects in Protoplasts Last at Least 96 h
We next determined what the onset of RNAi effects was, how long the RNAi effects would last, and how long protoplasts would stay alive in our assays. We incubated mock- and dsRNAPCS1-transfected protoplasts in W5 media and analyzed the abundance of PCS1 transcript by RT-PCR at specific time points. We determined that the abundance of PCS1 transcript in mock-transfected protoplasts remained constant (Fig. 4C). In contrast, RNAi effects in dsRNAPCS1-transfected protoplasts were observed within 24 h of transfection, and silencing effects persisted for at least 96 h (Fig. 4C). The approximately 90% decrease in PCS1 transcript abundance was observed after 72 h of transfection. After 96 h, however, due to the transient nature of dsRNA-induced RNAi in protoplasts, the abundance of PCS1 became more prominent but was still 75% lower in comparison with mock-transfected protoplasts (Fig. 4C).
These studies demonstrated that RNAi effects in protoplasts were triggered as early as 24 h post-transfection and lasted at least 96 h, thus allowing sufficient time for subsequent functional assays. From then on, we analyzed RNAi effects in protoplasts after 48 h of transfection, unless indicated otherwise.
Gene-Specific or Multiple Gene Interference of Expression of Arabidopsis Genes in Protoplasts by RNAi
Here, we sought to establish if transfecting dsRNA that targets sequences specific or shared among members of multigene family would effect expression of individual genes or several genes at the same time. To select a multigene family for these studies, we used the following criteria: (1) the multigene family must contain a relatively small number of members; (2) several genes of the multigene family must share a high degree of sequence similarity but also must be divergent from other family members; and (3) the expression levels of members of the gene subfamily must be well established.
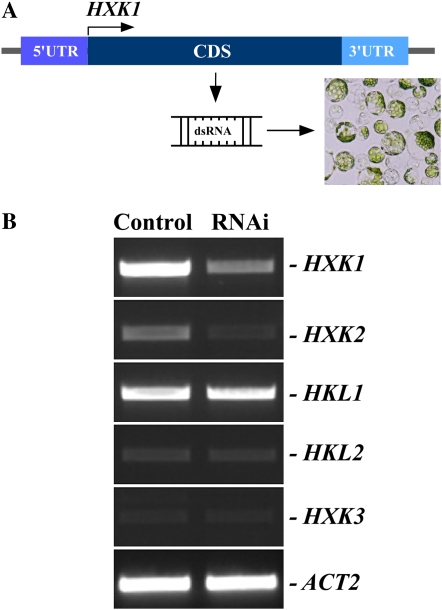
Using these criteria, we selected the hexokinase gene family in Arabidopsis, consisting of six members: HXK1, HXK2, and HXK3 (for Arabidopsis hexokinases 1, 2, and 3), and HKL1, HKL2, and HKL3 (Arabidopsis hexokinase-like proteins 1, 2, and 3; Karve et al., 2008). Among these, HXK2 shares 85% amino acid sequence identity with HXK1, whereas other family members are more divergent (Karve et al., 2008). These features of the HXK family members allowed us to assay the gene-specific and simultaneous interference with the expressions of HXK1 and HXK2 in protoplasts by targeting sequences that are unique or shared by these genes.
To silence HXK1 and HXK2 simultaneously, as a template for in vitro dsRNA synthesis we selected a part of the coding sequence of HXK1, which shares 83% identity with HXK2 but is distinct from other family members (Fig. 5A). RT-PCR analysis of transcript abundance of hexokinase gene family members from mock- and dsRNAHXK1-transfected protoplasts established that RNAi against sequences that are shared between HXK1 and HXK2 depleted the abundance of transcripts of both genes but did not affect expression of other family members (Fig. 5B). When abundance of HXK1 and HXK2 transcripts after RT-PCR analysis was normalized to the level of the actin gene, ACT2, a 54% and a 46% depletion, respectively, of corresponding transcripts was observed, while abundance of others remained at the level of the mock-transfected protoplasts (not shown). Because protoplasts were isolated primarily from Arabidopsis leaves, whereas HKL3 is expressed exclusively in Arabidopsis flowers (Karve et al., 2008), this gene was not amplified by RT-PCR and was not analyzed here. Our data indicated that we can silence genes simultaneously in protoplasts by targeting sequences that are shared among family members.
Figure 5.
Simultaneous interference with the expression of HXK1 and HXK2 in protoplasts by RNAi. A, dsRNA corresponding to the coding sequence of HXK1 was synthesized in vitro and delivered into protoplasts. Given the high sequence identity and similarity between HXK1 and HXK2 genes, dsRNA against HXK1 is also predicted to target HXK2. B, RT-PCR analysis of members of hexokinase gene family. Mock-transfected protoplasts (Control) and dsRNA-transfected protoplasts (RNAi) were collected for RT-PCR analysis after 24 h of transfection. ACT2 was amplified as a control for amount of a template. 5′-UTR and 3′-UTR are 5′- and 3′-end untranslated regions of HXK1, while CDS denotes the coding sequence of HXK1. [See online article for color version of this figure.]
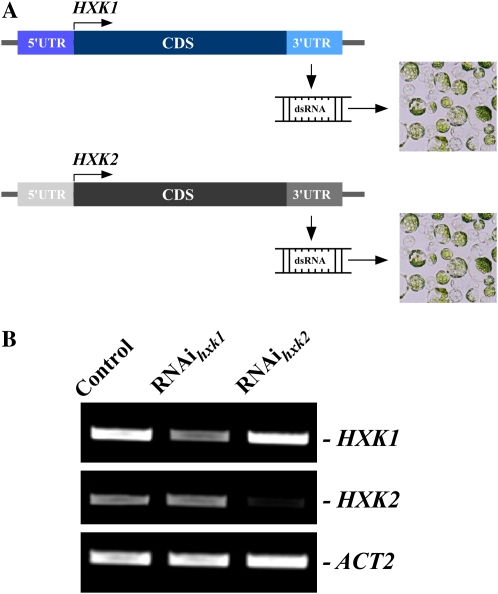
To test if RNAi against unique sequences of HXK1 and HXK2 would interfere with the expression of these genes in protoplasts individually, we selected their 3′-untranslated regions (UTRs) as templates for in vitro dsRNA synthesis (Fig. 6A). RT-PCR analysis of the abundance of HXK1 and HXK2 transcript in mock-, dsRNAUTRHXK1-, or dsRNAUTRHXK2-transfected protoplasts established that the gene-specific RNAi-mediated silencing can be achieved in protoplasts by targeting the gene-unique, 3′-UTR sequences (Fig. 6B). Transfection of protoplasts with dsRNAUTRHXK1 depleted the abundance of HXK1 but not of HXK2, whereas transfection of protoplasts with dsRNAUTRHXK2 depleted the abundance of HXK2 but not of HXK1 (Fig. 6B).
Figure 6.
Gene-specific interference with the expression of HXK1 and HXK2. A, dsRNAs corresponding to the 3′-UTR regions of HXK1 and HXK2 genes were synthesized in vitro and transfected into protoplasts to interfere with the expression of HXK1 or HXK2. B, RT-PCR analysis of HXK1 and HXK2 after RNAi against 3′-UTR regions of HXK1 or HXK2 genes. Mock-transfected protoplasts (Control) and dsRNA-transfected protoplasts (RNAihxk1 and RNAihxk2) were collected for RT-PCR analysis after 24 h of transfection. ACT2 was amplified as a control for amount of a template. [See online article for color version of this figure.]
Generation of a Single, PCR-Linked Template for dsRNA Synthesis Targeting of Two Genes Simultaneously
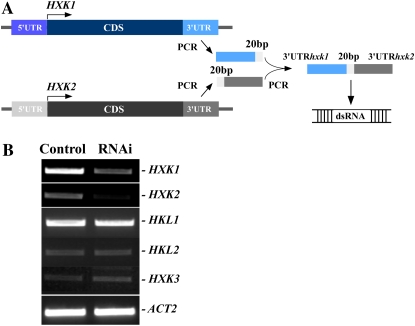
In some cases, for analysis of interactions of different pathways, it is necessary to silence several genes from these pathways simultaneously. This can be achieved by cotransfection of protoplasts with dsRNAs against 3′-UTR for the selected genes. However, cotransfection will result in mutual dilution of individual dsRNAs and will decrease individual dsRNA to protoplast ratios, and, because the gene silencing in protoplasts depends on the dose of transfected dsRNA (Fig. 4), will decrease the potency of RNAi-silencing effects. In addition, cotransfection of two or more dsRNAs imposes a difficulty in accessing the transfection efficiency of cotransfected dsRNAs. To circumvent this, we ligated 3′-UTR sequences by PCR to obtain a single, hybrid template for in vitro dsRNA synthesis (Fig. 7A). We used HXK1 and HXK2 genes as model genes, because we have already established that we can interfere with their expression simultaneously and individually (Fig. 6). We transfected protoplasts with 10 μg of the hybrid dsRNAUTRHXK1-UTRHXK2.
Figure 7.
Gene-specific and simultaneous interference with the expression of HXK1 and HXK2 using PCR-linked UTR regions as dsRNA templates. A, DNA fragments corresponding to 3′-UTR regions of HXK1 and HXK2 were PCR amplified, and, in the second round of PCR, were linked with a 20-bp linker (20 bp). The resulting PCR product encompassing 3′-UTR regions of both HXK1 and HXK2 genes was used as a template for in vitro dsRNA synthesis. B, RT-PCR analysis of the members of the HXK gene family after RNAi using the hybrid dsRNA, synthesized from a template described in A. Mock-transfected protoplasts (Control) and dsRNA-transfected protoplasts (RNAi) were collected for RT-PCR analysis after 24 h of transfection. [See online article for color version of this figure.]
RT-PCR analysis revealed that transfection of protoplasts with hybrid dsRNAUTRHXK1-UTRHXK2 decreased the abundance of HXK1 and HXK2 transcripts simultaneously (Fig. 7B). As expected, expression of other family members was not affected (Fig. 7B). Comparison of the level of the suppression of HXK1 and HXK2 expression by different sources of dsRNAs (Table III) demonstrated that the more potent RNAi silencing was achieved when protoplasts were transfected with the linked dsRNAUTRHXK1-UTRHXK2. When abundance of HXK1 and HXK2 transcripts were normalized to the level of ACT2, 1.3- and 2-fold more potent suppression of HXK1 and HXK2 expression, respectively, was observed when protoplasts were transfected with the hybrid dsRNAUTRHXK1-UTRHXK2 than when transfected with dsRNA against sequence that is shared between both family members or, in the case of HXK1, when transfected with dsRNA against HXK1 3′-UTR (Table I). These data show that transfection of a hybrid dsRNA against UTRs of several genes is a powerful approach for their simultaneous silencing.
Table III.
List of oligos that were used for RT-PCR analysis
| Genes | Directions | Oligos (5′–3′) |
|---|---|---|
| PCS1 | Forward | GCTGGATCGAAATAGCCAAG |
| Reverse | GCTTCAGGACCACATTCACA | |
| ECS1 | Forward | ATTGCCTATCTTGCCTCTGG |
| Reverse | ATCCGTTTGGCTTTCCTTCT | |
| HXK1 | Forward | GGACGTGTTTTGGCTATCCTC |
| Reverse | GAAATCCAGCGTGTGATCAA | |
| HXK2 | Forward | GACAGAGTACGACCACTCTCTAGATG |
| Reverse | TTAACTTGTTTCAGAGTCATCTTCAAG | |
| HKL1 | Forward | AGGAAAACGGGTCCGATTCA |
| Reverse | TGCTTCTCCCGCTCGTGAT | |
| HKL2 | Forward | GGGCTTTATCCTTTGGACATTT |
| Reverse | TCATACGGATGGTATTGTTTGAAC | |
| HXK3 | Forward | CCAATTACATCGTGATGTCCG |
| Reverse | GCGGAAGAATCTTAGAGAACCC | |
| HKL3 | Forward | GAACTATTACTGTTACGTGGGACG |
| Reverse | CAGTCTCCCGTTCTTCGGAT |
Table I.
Comparison of the relative abundance of transcripts in protoplasts after RNAi using different DNA templates
Relative abundance of transcripts after RT-PCR was normalized to the level of ACT2, encoding actin, using GelQuant software.
| DNA Templates for dsRNA Synthesis | Size of dsRNA | Depletion of Relative Transcript Abundance after RNAi
|
|
|---|---|---|---|
| HXK1 | HXK2 | ||
| bp | % | ||
| cDNA of HXK1 | 524 | 53 | 46 |
| 3′-UTR of HXK1 | 281 | 55 | 0 |
| 3′-UTR of HXK2 | 207 | 0 | 92 |
| 3′-UTR of HXK1 + 3′-UTR of HXK2 | 500 | 73 | 92 |
RNAi in Protoplasts Is Suitable for Functional Analysis of Plant Genes
The studies described above have established that the specific or simultaneous depletion of gene transcripts is successfully achieved in protoplasts by transfection of in vitro-synthesized dsRNAs. We also demonstrated that RNAi effects lasted at least 96 h. However, is RNAi depletion of transcripts of endogenous genes accompanied by a decrease in a polypeptide level? Can we use RNAi in protoplasts to study the function of silenced genes? Are RNAi phenotypes in protoplasts comparable with those observed in genetic mutants? To address these questions, we selected an RNAi-targeted gene using the following criteria: (1) the RNAi-targeted gene should be a single-copy gene with the established expression level; (2) antibody for the analysis of the corresponding polypeptide by western-blot analysis should be available; (3) tests for analysis of the functional activity of the encoded polypeptide should be well established and simple; and (4) mutant alleles with well-characterized phenotypes should be available.
Using these criteria, we selected ECS1 (alias cad2) encoding β-glutamylcysteine synthase (β-ECS) as an RNAi target. ECS1 is a key enzyme in biosynthesis of the ubiquitous tripeptide GSH (β-Gly-Cys-Gly). Therefore, its functional activity in RNAi protoplasts can be evaluated by assaying their GSH content (Cobbett et al., 1998). In addition, the expression of ECS1 has been established, antibodies are commercially available, and the cad2-1 mutant allele of ECS1 has been generated (Cobbett et al., 1998). It has also been determined that the decrease in the activity of ECS1 of cad2-1 is accompanied by a 55% decrease in cellular GSH content (Cobbett et al., 1998).
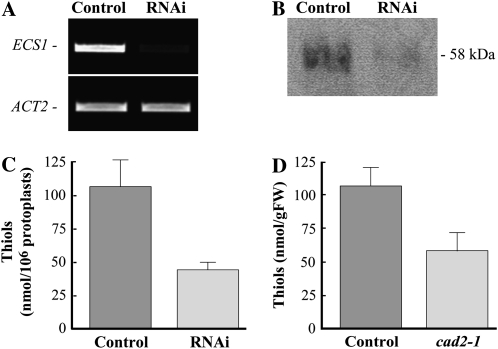
Because ECS1 is a single-copy gene, we selected 548 bp of its coding sequence as an RNAi target, PCR amplified it from the Arabidopsis cDNA library, synthesized dsRNAECS1 in vitro, and transfected it into protoplasts. RT-PCR analysis of mock- and dsRNAECS1-transfected protoplasts after 48 h of transfection established that the abundance of ECS1 transcript is substantially decreased in RNAi, but not in control, protoplasts (Fig. 8A).
Figure 8.
RNAi silencing of ECS1 in protoplasts interferes with the expression of ECS1 RNA and polypeptide, and decreases the ability of protoplasts to accumulate GSH. A, RT-PCR analysis of the abundance of ECS1 in protoplasts transfected with dsRNAECS1 (RNAi) or water (Control). B, SDS-PAGE and western-blot analysis of ECS1 polypeptide in mock-transfected (Control) or RNAi protoplasts (RNAi). C, RP-HPLC analysis of GSH content in protoplasts transfected with water (Control) or with dsRNAECS1 (RNAi). Protoplasts were collected and analyzed after 48 h of transfection. D, RP-HPLC analysis of GSH content in wild-type Arabidopsis (Control) and cad2-1 mutants (cad2-1). FW, Fresh weight.
Importantly, RNAi in protoplasts not only depleted the abundance of ECS1 transcript but substantially decreased the level of the corresponding polypeptide (Fig. 8B). As determined by SDS/PAGE and western-blot analysis, a single polypeptide species that reacted with the rabbit polyclonal anti-ECS1 antibody was presented in lysates from control and dsRNAECS1-transfected protoplasts. However, in comparison with control protoplasts, the level of the polyclonal anti-ECS1 antibody-reactive polypeptide was 3-fold lower in dsRNAECS1-transfected protoplasts (Fig. 8B).
We then asked if RNAi against ECS1 would affect the GSH content of protoplasts. Reverse-phase HPLC (RP-HPLC) analysis of nonprotein thiol compounds in mock-transfected protoplasts revealed a peak of GSH, the chromatographic properties of which were indistinguishable from commercially available GSH standard (Sigma-Aldrich). The aggregate GSH content in control protoplasts was 106.9 ± 19.9 nmol/106 protoplasts (Fig. 8C). The GSH content of RNAi protoplasts was 2.5-fold lower and was consistent with the fold-decrease of ECS1 polypeptide (Fig. 8, B and C). These data showed that transfection of dsRNA into protoplasts not only affected the transcript level but also the abundance of the translated polypeptide and, as judged by the decrease in accumulation of the end product of the encoded enzymatic activity of the RNAi targeted gene, interfered with the gene's function.
Importantly, the fold-decrease in the GSH content resulting from RNAi silencing of ECS1 in protoplasts is comparable with that observed in cad2-1 mutants bearing a 6-bp deletion in the sixth exon of the ECS1 gene (Cobbett et al., 1998). The content of GSH in cell-free extracts from cad2-1 mutants was 1.8-fold lower than in extracts from wild-type Arabidopsis (Fig. 8D). Taken together, our data demonstrate that RNAi can be used as a versatile approach to study gene function in protoplasts.
CONCLUSION AND FUTURE PERSPECTIVES
In this manuscript, we presented data demonstrating that RNAi can be effectively used for functional analysis of genes in protoplasts. We substantiated the results of An et al. (2003, 2005) by presenting the following findings. First, RNAi-mediated silencing was induced in protoplasts by transfecting in vitro-synthesized dsRNA. This circumvented the necessity to clone dsRNA into expression vectors, which can be time consuming and labor intensive, especially for the genome-wide studies. Second, RNAi effects in protoplasts depended on a dose of transfected dsRNA. Third, RNAi effects in protoplasts lasted at least 96 h, allowing sufficient time for subsequent functional analysis. Fourth, RNAi in protoplasts silenced individual genes as well as multiple members of gene families simultaneously. Fifth, transfecting protoplasts with dsRNAs possessing linked 3′-UTRs of two genes that are members of a multigene subfamily silenced both genes simultaneously, but did not interfere with the expression of other family members. In doing so, we circumvented the necessity to cotransfect protoplasts with two dsRNAs against targeted genes, avoided the inevitable mutual dilution of dsRNAs in the transfection media, a possibility of different transfection efficiencies of two dsRNAs, and thus achieved potent gene silencing of both genes simultaneously. We expect that this approach will be especially valuable for the analysis of interactions among distinct pathways when it will be necessary to silence genes from two (or more) families that share high sequence similarity. Sixth, transfection of dsRNA depleted the abundance not only of the transcript of a targeted gene but also of the corresponding polypeptide. This was a significant finding, because protoplasts are nongrowing cells that have a limited lifespan. Therefore, if the studied gene is not induced under experimental condition, RNAi silencing of its function in protoplasts will be contingent on the turnover rates of the corresponding polypeptide. In the case of ECS1, we achieved a 72% decrease in the level of polypeptide after 48 h of protoplast transfection with dsRNAECS1, whereas in our assays, protoplasts were viable, and RNAi-depletion of a gene transcript lasted even longer, for 96 h. Therefore, this method should be appropriate for functional studies of even relatively stable proteins and is particularly appropriate for studies of conditionally inducible genes. Seventh, transient RNAi knockdown of ECS1 in protoplasts led to a loss-of-ECS1 function: protoplasts transfected with dsRNAECS1 accumulated 60% less of the end product of ECS1 enzymatic activity, GSH. Eighth, RNAi in protoplasts yields results comparable with those of genetic mutants for the same gene. Ninth, we adapted protoplast isolation protocol to 14-d-old seedlings. Therefore, the described procedure is rapid; 20 d are sufficient for analysis. This time includes growing Arabidopsis, isolation of protoplasts, in vitro dsRNA synthesis, transfection of dsRNA into protoplasts, and analysis of phenotypes. Taking together, RNAi can silence gene functions in protoplasts, providing a rapid and a cost-efficient tool for reverse-genetic studies.
Limitations of using RNAi in protoplasts for functional analysis of genes include potential artifacts that might emerge due to the removal of plant cell walls, disruption of cell-to-cell communications, and the artificial protoplast culture conditions. Clearly, phenotypes observed in RNAi protoplasts have to be verified and complemented by studies in planta. Nevertheless, RNAi in protoplasts can be used as a rapid and affordable initial screen for selection of candidate genes for subsequent studies in planta using available resources such as transposon/T-DNA insertion lines or RNAi transgenic plants. While it is unlikely that RNAi in protoplasts will be appropriate for studies of genes involved in developmental processes, it presents an excellent approach for biochemical characterization of proteins with enzymatic activities (e.g. ECS1; Fig. 8), with many of them having no experimentally assigned functions. This method is particularly suitable for deciphering components of metabolic pathways and establishing of metabolic networks and, perhaps, can be adapted to high-throughput metabolomics. Because protoplasts can be isolated from different sources, this approach can be used to study tissue-specific biochemical processes in Arabidopsis and other plant species.
RNAi in protoplasts can be also used as an initial step for identifying genes required for a particular function. For instance, this approach is particularly valuable if gene silencing is anticipated to result in a conditional lethal phenotype (e.g as for genes required for tolerance to xenobiotics, cold, heat). In this case, combining protoplast viability assays using fluorescent vital dyes (e.g. propidium iodide, SYTOX Orange, etc.) with the fluorescence-activated cell sorting analyses will provide a powerful and rapid approach for selecting gene candidates required for tolerance to applied stress.
Another interesting example involves cell wall metabolism. It is well documented in different plant species that protoplasts regenerate their cell walls (Burgess and Fleming, 1974; Wenck and Marton, 1995); hence, RNAi in protoplasts will greatly assist in discovery and analyses of genes involved in cell wall biogenesis.
To conclude, RNAi in protoplasts complements existing approaches for functional analyses of genes as it provides the time-efficient and affordable initial step for selecting of genes of interest for further in depth studies in planta. The recent advances in sequencing of many plant genomes provide information for targeted and systematic screens of genes and gene families, awaiting development of novel functional genetic approaches, such as presented here. Therefore, adopting RNAi in protoplasts to a high-throughput, genome-wide phenotyping technology will greatly facilitate assigning functions to unknown genes not only in Arabidopsis but also in other plant species.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) Columbia ecotype seeds were sterilized in 70% ethanol, 1.8% bleach solution (made up by diluting a household Clorox containing 6% sodium hypochlorite), and 0.1% Tween 20. Seeds were sown on 1/2 Murashige and Skoog medium, pH 5.7, adjusted with 1 n KOH, supplemented with 1% Suc and 0.7% agar and then kept for 24 h at 4°C in the dark for stratification. Seedlings were germinated and grown in an 8-h-light/16-h-dark photoperiod (at a photosynthetic photon flux density of 250 μmol m−2 s−1) at a 23°/19°C light/dark temperature regime and 75% relative humidity.
Isolation of Protoplasts from Arabidopsis Seedlings
Protoplasts were isolated from Arabidopsis seedlings using a modified method of Chen and Halkier (2000). Briefly, 2 g of 14-d-old Arabidopsis seedlings were sliced with a razor blade to 1-mm strips in 15 mL of filter-sterilized TVL (0.3 m sorbitol, 50 mm CaCl2) solution. Twenty milliliters of enzyme solution (0.5 m Suc, 10 mm MES-KOH, pH 5.7, 20 mm CaCl2, 40 mm KCl, 1% Cellulase [Onozuka R-10], 1% Macerozyme [R10]) were added to the sliced seedlings, and the mixture was agitated at 35 rpm in the dark at room temperature. After 16 to 18 h, the released protoplasts were sieved through eight layers of cheesecloth, pre-wet in W5 solution (0.1% [w/v] Glc, 0.08% [w/v] KCl, 0.9% [w/v] NaCl, 1.84% [w/v] CaCl2, 2 mm MES-KOH, pH 5.7) and transferred into 50-mL Falcon centrifuge tubes. Protoplasts and plant debris retained on the cheesecloth were gently sieved-through one more time by washing the cloth with 15 mL of W5 solution. Sieved-through protoplasts were combined in a 50-mL Falcon centrifuge tube, overlaid with 10 mL of W5 solution, and centrifuged for 7 min at 100g. Protoplasts were collected at the interface of enzyme and W5 solutions (Fig. 1A). The protoplast yield was evaluated by cell counting with a hemocytometer. Using this procedure, we typically harvested 5 to 10 × 106 protoplasts from 1 g of fresh seedlings.
In Vitro Synthesis of dsRNA
DNA templates were synthesized by PCR from Arabidopsis cDNA and engineered to contain the minimal T7-RNA polymerase promoter sequence (TAATACGACTCACTATAGGG) on both 5' and 3' ends (Sastry and Ross, 1997). Primers used to amplify DNA of targeted genes are described in Table II. The PCR conditions were as follows: denaturing step at 94°C for 3 min followed by 30 cycles of 94°C for 45 s, 57°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 10 min. dsRNAs were synthesized in vitro using the Ambion MEGAscript T7 kit (Ambion) according to the manufacturer's recommendation with the exception that in vitro transcription was allowed to proceed for at least 6 h. DNA templates were removed with the RNases-free DNase (Invitrogen), dsRNAs were purified with the RNAeasy kit (Qiagen), reconstituted with deionized, sterile water, and quantified using UV spectrophotometry. The standard output of dsRNA in a 20-μL reaction was between 100 and 200 μg.
Table II.
List of oligos that were used to amplify DNA templates for in vitro dsRNA synthesis
| Size of dsRNA | Targeted Gene | Directions | Oligos (5′–3′) |
|---|---|---|---|
| bp | |||
| 547 | PCS1 | Forward | GCGCCTAATACGACTCACTATAGGGAGAGAACCTCTGGAAGTAGTGAAGGAA |
| Reverse | GCGCCTAATACGACTCACTATAGGGAGACTGTGAACTTACAAGACGAGGAAC | ||
| 540 | EGFP | Forward | GCGCCTAATACGACTCACTATAGGGAGAGCCAACACTTGTCACTACTTTCTC |
| Reverse | GCGCCTAATACGACTCACTATAGGGAGACATGTGGTCTCTCTTTTCGTTG | ||
| 548 | ECS1 | Forward | TAATACGACTCACTATAGGGAGATCATGCCAAAGGGGAGATAC |
| Reverse | TAATACGACTCACTATAGGGAGACTCTTCAACCGAACCTCTGG | ||
| 524 | HXK1 (cDNA) | Forward | AAGCTTTAATACGACTCACTATAGGGCTGCTTCCAAAATCAGGAGAAA |
| Reverse | GGATCCTAATACGACTCACTATAGGGGTAATGCTCAAACAATCCACCA | ||
| 281 | HXK1 (3′-UTR) | Forward | GCGCCTAATACGACTCACTATAGGGAGATCGCTATCAGAAAACGCCTAA |
| Reverse | GCGCCTAATACGACTCACTATAGGGAGACTCCAGTGAAGTGAGCTTTGA | ||
| 207 | HXK2 (3′-UTR) | Forward | GCGCCTAATACGACTCACTATAGGGAGATTGTTTTGCCGTTAGGGT |
| Reverse | GCGCCTAATACGACTCACTATAGGGAGAACAACATTGGCTGGTCGTTT | ||
| HXK1 (3′-UTR) + Linker | Forward | GCGCCTAATACGACTCACTATAGGGAGATCGCTATCAGAAAACGCCTAA | |
| Reverse | GGACCGTCACAATAGTACATAGAGAGGCCTCCAGTGAAGTGAGCTTTGA | ||
| HXK2 (3′-UTR) + Linker | Forward | GCCTCTCTATGTACTATTGTGACGGTCCTTGTTTTGCCGTTAGGGTTT | |
| Reverse | GCGCCTAATACGACTCACTATAGGGAGAACAACATTGGCTGGTCGTTT |
Transfection of Protoplasts with Plasmid DNA or dsRNAs
Protoplasts were transfected using a procedure adopted from Yoo et al. (2007). Briefly, protoplasts collected at interface of the enzyme solution and the W5 solution and were washed three times by reconstituting in W5 solution and pelleting at 100g for 2 min. After the third wash, protoplasts were resuspended in MMG solution (4 mm MES-KOH, pH5.7, 0.4 m mannitol, 15 mm MgCl2) to reach a final concentration of 5 to 10×105 protoplasts/mL. Aliquots of protoplasts (100 μL; 0.5–1.0×105 protoplasts/mL) were transferred into a 2-mL round-bottom microcentrifuge tube and mixed gently with the plasmid DNA (6 μg in 10 μL) or with dsRNA (1, 5, or 10 μg in 10 μL). In control transfections, we omitted DNA or RNA and instead used equivalent volumes of deionized, sterile water (mock-transfection). Transfection was initiated by the addition of 110 μL of PEG-calcium solution (40% PEG-4000, 0.2 m mannitol, 100 mm CaCl2). Protoplasts were mixed with PEG-calcium solution by gently tapping the tube and incubating for 5 min at room temperature. Transfection was terminated by diluting the mixture with 600 μL of W5 solution. Transfected protoplasts were collected by centrifugation for 2 min at 100g, resuspended in 1 mL of W5 solution, and kept in the dark for the time periods indicated in the figures prior to further analyses.
RT-PCR Analysis of Gene Expression in RNAi Protoplasts
RNAi and control protoplasts (105 cells/mL) were collected by centrifugation for 2 min at 100g. Total RNA was extracted with TRIzol reagent (Invitrogen, 1 mL/105 protoplasts) following the manufacturer's protocol. RNA aliquots (10 μL) were subjected to RT with M-MuLV Reverse Transcriptase (New England Biolabs) followed by PCR analysis using GoTag Green Master mix (Promega). PCR conditions were as follows: denaturing at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 60 s, and a final extension at 72°C for 5 min. Primers that were used for RT-PCR analyses are described in Table III. The effectiveness of RNAi was evaluated by analysis of aliquots from RT-PCR reactions on a 1.2% gel and comparing the relative abundance of transcripts of RNAi-targeted genes to the level of the Arabidopsis actin gene, ACT2, using the GelQuant software, version 2.7 (DNR Bio-Imaging Systems).
Western-Blot Analysis of ECS1 in RNAi Protoplasts
RNAi and control protoplasts (105 cells/mL) were collected by centrifugation at 100g for 2 min and lysed by resuspension in 10 μL of the Laemmli buffer. Protoplast proteins were denatured at 50°C for 10 min, separated by SDS-PAGE on 10% (w/v) gels, and electrotransferred in Towbin buffer (Towbin et al., 1979) to a nitrocellulose filter. The nitrocellulose blots were probed with the primary rabbit polyclonal anti-ECS antibody (1:1,000 dilution; Agrisera) and with the secondary HP-conjugated anti-rabbit antibody (1:2,500; GE Healthcare). Immunoreactive bands were visualized with LumiGLO system (KPL).
Analysis of GSH Content
GSH content was analyzed in RNAi and control protoplasts and in plant extracts by a combination of RP-HPLC and thiol quantitation with Ellman's reagent as described (Vatamaniuk et al., 2000). Aliquots of protoplasts corresponding to 106 cells or cell-free plant extracts (10–20 μg of protein) were made 5% (w/v) with 5-sulfosalicylic acid and centrifuged before aliquots of the supernatant (50 μL) were loaded onto an Econosphere C18, 150 × 4.6-mm RP-HPLC column (Alltech). The column was developed with a linear gradient of water, 0.05% (v/v) phosphoric acid, and 17% (v/v) acetonitrile, 0.05% (v/v) phosphoric acid at a flow rate of 1 mL/min. For the quantitation of GSH, thiols were estimated spectrophotometrically at 412 nm by reacting aliquots (500 μL) of the column fractions with 0.8 mm 5,5′-dithiobis(2-nitrobenzoic acid) (500 μL) dissolved in 250 mm phosphate buffer, pH 7.6 (Ellman, 1959). Commercially available GSH (Sigma) was used as a RP-HPLC standard and for calibration.
Accession numbers for genes used in this study (accession numbers are in parenthesis) are as follows: PCS1 (At5g44070), ECS1 (At4g23100), HXK1 (At4g29130), HXK2 (At2g19820), HKL1 (At1g50460), HKL2 (At3g20040), HXK3 (At1g47840), HKL3 (At4g37840), and ACT2 (AT3G18780).
Acknowledgments
We thank Dr. Jian Hua, Cornell University, for the pSAT vector; Dr. Wolf Frommer, Carnegie Institution, Stanford University, for providing primers for PCR amplification and RT-PCR analysis of HXK family members; Dr. Chris Cobbett, University of Melbourne, Australia, for providing seeds of cad1-3 mutants of Arabidopsis; and Dr. Sixue Chen, University of Florida, for advice on isolating protoplasts. We thank Andy Chen, Dr. Tim Setter, and Dr. Leon Kochian, Cornell University, and Dr. Jose Alonso and Dr. Anna Stepanova, North Carolina State University, for constructive discussions on the manuscript.
This work was supported by the Cornell University Agricultural Experiment Station (Hatch Grant no. NYC–125433 and Cornell Start-Up Grant to O.K.V.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Olena K. Vatamaniuk (okv2@cornell.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open access articles can be viewed online without a subscription.
References
- Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22 1559–1566 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- An CI, Sawada A, Fukusaki E, Kobayashi A (2003) A transient RNA interference assay system using Arabidopsis protoplasts. Biosci Biotechnol Biochem 67 2674–2677 [DOI] [PubMed] [Google Scholar]
- An CI, Sawada A, Kawaguchi Y, Fukusaki E, Kobayashi A (2005) Transient RNAi induction against endogenous genes in Arabidopsis protoplasts using in vitro-prepared double-stranded RNA. Biosci Biotechnol Biochem 69 415–418 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genomics Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bai J, Binari R, Ni JQ, Vijayakanthan M, Li HS, Perrimon N (2008) RNA interference screening in Drosophila primary cells for genes involved in muscle assembly and maintenance. Development 135 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431 356–363 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366 [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS (2005) An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol (Stuttg) 7 251–257 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HFA, Paro R, Perrimon N (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303 832–835 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22 268–280 [DOI] [PubMed] [Google Scholar]
- Burgess J, Fleming EN (1974) Ultrastructural observations of cell wall regeneration around isolated tobacco protoplasts. J Cell Sci 14 439–449 [DOI] [PubMed] [Google Scholar]
- Chen S, Halkier BA (2000) Characterization of glucosinolate uptake by leaf protoplasts of Brassica napus. J Biol Chem 275 22955–22960 [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA 97 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C, Howden R, Rolls B (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1 of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J 16 73–78 [DOI] [PubMed] [Google Scholar]
- Cocking EC (1960) A method for the isolation of plant protoplasts and vacuoles. Nature 187 962–963 [Google Scholar]
- Ellman GL (1959) Tissue sulfhydryl groups. Arch Biochem Biophys 82 70–77 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811 [DOI] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS (2005) A systematic RNAi screen for longevity genes in C. elegans. Genes Dev 19 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Waterhouse PM, David R, Engelke AJJR (2005) Constructs and methods for hairpin RNA-mediated gene silencing in plants. In DR Engelke, JJ Rossi, eds, Methods in Enzymology, Vol 392. Academic Press, San Diego, pp 24–35 [DOI] [PubMed]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237 [DOI] [PubMed] [Google Scholar]
- Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD (2008) Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 1017–1026 [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry SS, Ross BM (1997) Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J Biol Chem 272 8644–8652 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Gene expression: total silencing by intron-spliced hairpin RNAs. Nature 407 319–320 [DOI] [PubMed] [Google Scholar]
- Stam M, de Bruin R, Kenter S, van der Hoorn RAL, van Blokland R, Mol JNM, Kooter JM (1997) Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J 12 63–82 [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travella S, Klimm TE, Keller B (2006) RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol 142 6–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA (1999) AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc Natl Acad Sci USA 96 7110–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA (2000) Mechanism of heavy metal ion activation of phytochelatin (PC) synthase: blocked thiols are sufficient for PC synthase-catalyzed transpeptidation of glutathione and related thiol peptides. J Biol Chem 275 31451–31459 [DOI] [PubMed] [Google Scholar]
- Vidali L, Augustine RC, Kleinman KP, Bezanilla M (2007) Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19 3705–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4 29–38 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411 834–842 [DOI] [PubMed] [Google Scholar]
- Wenck AR, Marton L (1995) Large-scale protoplast isolation and regeneration of Arabidopsis thaliana. Biotechniques 18 640–643 [PubMed] [Google Scholar]
- Wilson JA, Richardson CD (2003) Induction of RNA interference using short interfering RNA expression vectors in cell culture and animal systems. Curr Opin Mol Ther 5 389–396 [PubMed] [Google Scholar]
- Worby CA, Simonson-Leff N, Dixon JE (2001) RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci STKE 2001 pl1. [DOI] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocols 2 1565–1572 [DOI] [PubMed] [Google Scholar]