Abstract
The role of the Psb28 protein in the structure and function of the photosystem II (PSII) complex has been studied in the cyanobacterium Synechocystis sp. PCC 6803. The protein was localized in the membrane fraction and, whereas most of the protein was detected as an unassembled protein, a small portion was found in the PSII core complex lacking the CP43 antenna (RC47). The association of Psb28 with RC47 was further confirmed by preferential isolation of RC47 from the strain containing a histidine-tagged derivative of Psb28 using nickel-affinity chromatography. However, the affinity-purified fraction also contained a small amount of the unassembled PSII inner antenna CP47 bound to Psb28-histidine, indicating a structural relationship between Psb28 and CP47. A psb28 deletion mutant exhibited slower autotrophic growth than wild type, although the absence of Psb28 did not affect the functional properties of PSII. The mutant showed accelerated turnover of the D1 protein, faster PSII repair, and a decrease in the cellular content of PSI. Radioactive labeling revealed a limitation in the synthesis of both CP47 and the PSI subunits PsaA/PsaB in the absence of Psb28. The mutant cells contained a high level of magnesium protoporphyrin IX methylester, a decreased level of protochlorophyllide, and released large quantities of protoporphyrin IX into the medium, indicating inhibition of chlorophyll (Chl) biosynthesis at the cyclization step yielding the isocyclic ring E. Overall, our results show the importance of Psb28 for synthesis of Chls and/or apoproteins of Chl-binding proteins CP47 and PsaA/PsaB.
PSII is a multisubunit pigment-protein complex of plants, algae, and cyanobacteria, which is responsible for oxidation of water and reduction of plastoquinone during oxygenic photosynthesis (Barber, 2006). In the heart of the complex, there are two similar membrane-spanning proteins, D1 and D2, that bind the cofactors involved in primary charge separation (Nanba and Satoh, 1987) and subsequent electron transfer within PSII (for review, see Barber, 2006). Peripherally to the D1-D2 heterodimer, there are two chlorophyll (Chl)-binding inner antenna proteins, CP47 and CP43, that deliver energy to the reaction center (RC), driving electron transfer. In addition, CP43 also provides important ligands to the Mn4Ca cluster, the site of water oxidation (Ferreira et al., 2004; Loll et al., 2005). These four large proteins are surrounded by a number of smaller membrane polypeptides (for review, see Shi and Schröder, 2004). One of them, the so-called PsbW, was originally detected in the isolated RC complex from spinach (Spinacia oleracea; Irrgang et al., 1995; Lorković et al., 1995). The mature protein with a predicted one-transmembrane α-helix in the central hydrophobic region seems to have (unlike most of PSII membrane proteins) the N terminus oriented into the lumen in close vicinity to the extrinsic, nuclear-encoded 33-kD PsbO protein. Cross-linking experiments also indicated a close association of PsbW with D1, D2, and the α-subunit of cytochrome (cyt) b-559 in the isolated RC complex (Irrgang et al., 1995; Lorković et al., 1995). At variance with these results, Rokka et al. (2005) located PsbW predominantly in PSII-light-harvesting complex II (LHCII) supercomplexes and only minor amounts were found in PSII core dimers and monomers. In transgenic plants of Arabidopsis (Arabidopsis thaliana) lacking the PsbW protein, the stability of the dimeric PSII was diminished and the PSII-LHCII supercomplexes could not be detected. It has been suggested that PsbW functions as a linker for LHCII binding to the PSII complex (Shi et al., 2000). Because LHCII is absent in cyanobacteria, it was intelligible that the PsbW was not detected in these oxygenic autotrophs. Nevertheless, N-terminal sequencing and mass spectrometric analyses of protein subunits in the purified His-tagged PSII from Synechocystis sp. PCC 6803 (Synechocystis 6803) revealed the presence of an unknown protein with 16% sequence identity to PsbW from Arabidopsis (Kashino et al., 2002). This protein was designated as Psb28 (also Psb13 or ycf79). Its amino acid sequence suggests that it is a rather hydrophilic protein without a transmembrane helix and is larger than PsbW (about 13 kD). In the recent crystal structures of the cyanobacterial PSII (Ferreira et al., 2004; Loll et al., 2005), this protein was not identified and it remains an issue of contention whether the protein is a true PSII subunit, a transiently associated assembly factor, or just an impurity of the preparation. The relatively low content of this protein in the isolated preparation suggested that the two latter possibilities are more probable. Very recently, the protein has been detected as a component of PSII complexes in Synechocystis depleted of phosphatidylglycerol (Sakurai et al., 2007). It has been proposed that the protein may play a regulatory role during the assembly of PSII. A gene encoding a similar soluble protein has also been found in the genome of Arabidopsis and the protein was designated PsbW-like.
Here, we present a detailed analysis of the role of Psb28 in the structure and function of PSII in Synechocystis 6803. The results showed that Psb28 is not a component of the fully assembled dimeric PSII core complex, but it is preferentially bound to PSII assembly intermediates containing the inner antenna CP47. The results support the role of the protein in biogenesis of certain Chl-binding proteins via regulating synthesis of their apoproteins or Chls.
RESULTS
Psb28 Protein Is Associated with the PSII Assembly Complexes Containing CP47
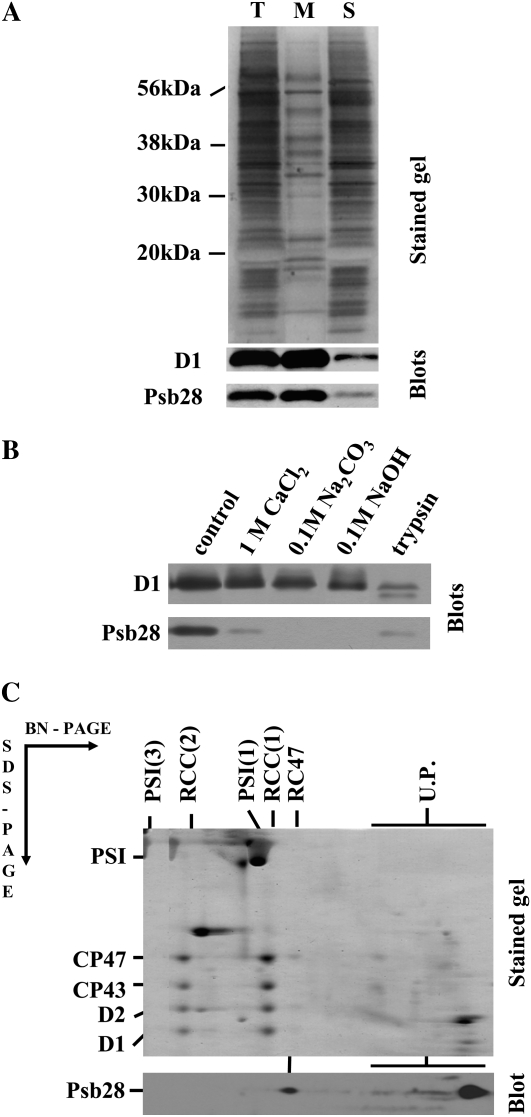
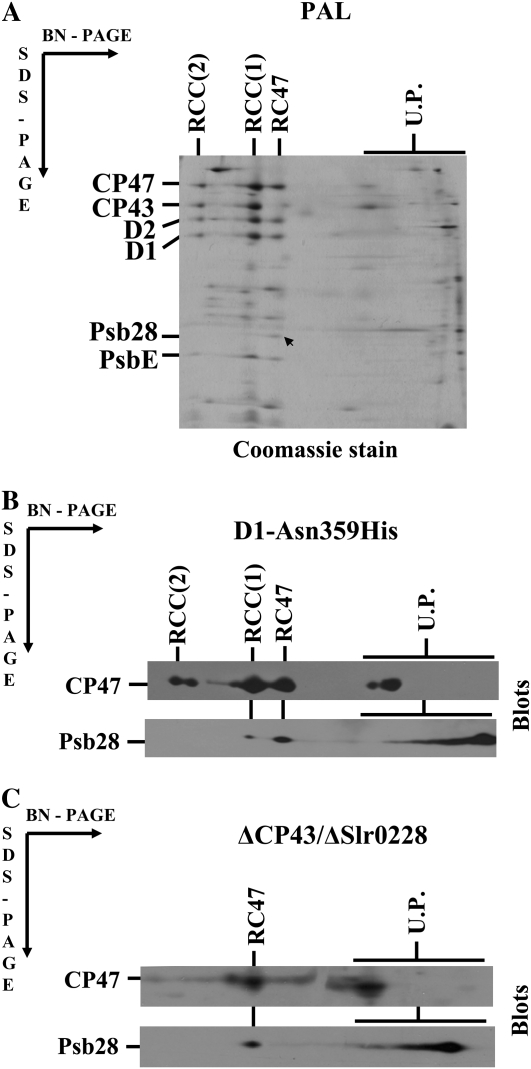
Psb28 was previously identified as a minor component of PSII complexes purified using nickel-affinity chromatography from the strain expressing His-tagged CP47 (Kashino et al., 2002). To provide more rigorous information about the localization of Psb28, we first screened for its presence in the membrane-associated and soluble protein fractions of a Synechocystis whole-cell extract using specific antibodies raised against the last 15 amino acid residues of the protein (Fig. 1A). The vast majority of the protein was found in the membrane fraction. However, the protein was only loosely bound as documented by removal of >90% of the protein by treatment of the membranes with 1 m CaCl2. Furthermore, the protein was completely washed out from membranes by 0.1 m sodium carbonate or 0.1 m sodium hydroxide. Ninety percent of the protein was also removed by treatment with trypsin, which acts only on the cytoplasmic side of this type of membrane preparation, cleaving the D1 protein into specific fragments (Komenda et al., 2002). Thus, Psb28 is the protein peripherally associated with the cytoplasmic side of the membranes. To identify the membrane-binding site of Psb28, we analyzed membrane protein complexes of wild type using two-dimensional (2D) separation of membrane proteins consisting of blue native (BN)-PAGE in one direction and denaturing PAGE in the second direction (2D BN/SDS-PAGE). The majority (80%–90%) of Psb28 was detected at the end of the BN gel in the low-molecular-weight region, indicating that it is not assembled into any large membrane complex (Fig. 1C, blot). However, a small portion of the protein was found in the position corresponding to the migration of a PSII core subcomplex lacking the inner antenna CP43 (termed RC47), while the dimeric and monomeric core complexes RCC(1) and RCC(2) were seemingly free of this subunit. Because the wild type contains only a small amount of RC47, Psb28 bound to this complex was almost undetectable. Therefore, we also analyzed PSII complexes in the phycobiliprotein-free strain called PAL (Ajlani and Vernotte, 1998), in the site-directed mutant D1-Asn-359His (Kuviková et al., 2005), and in the psbC/slr0228 double-deletion mutant ΔCP43/ΔSlr0228 (Komenda et al., 2006). PAL and D1-Asn-359His are autotrophic mutants with a high RC47:RCC ratio and the nonautotrophic ΔCP43/ΔSlr0228 deletion mutant accumulates a high level of RC47 as the only PSII complex due to the missing genes for CP43 and a FtsH protease involved in regulating the level of PSII assembly intermediates (Komenda et al., 2006). 2D analysis confirmed the presence of Psb28 in RC47 of PAL and the protein was apparent even in the stained gel (Fig. 2A, arrow). Its identity was verified by de novo sequencing of the band using mass spectrometry. In the site-directed mutant D1-Asn-359His, western blotting confirmed the presence of the protein in RC47, but a small amount of Psb28 was also found in the monomeric core complex RCC(1) (Fig. 2B). The Psb28 protein was also clearly detected in RC47 of the ΔCP43/ΔSlr0228 mutant (Fig. 2C). In both latter mutants, the band of unassembled Psb28 exhibited typical smearing approximately starting in the gel region in which the unassembled CP47 is located. This finding raised the question of whether these two proteins might form an unstable complex that is released during native electrophoresis (see below).
Figure 1.
Localization of the Psb28 protein in the membrane fraction of the Synechocystis proteins. A, The total cell extract from the wild-type strain after cell breakage (T), its membrane fraction after centrifugation (M), and supernatant after centrifugation (S) were analyzed for the presence of the D1 and Psb28 proteins by SDS-PAGE. The gel was either stained with Coomassie Blue (stained gel) or blotted to a PVDF membrane and D1 and Psb28 were detected by specific antibodies (blots).The amount of protein loaded for each sample corresponded to the number of cells containing 1 μg of Chl. B, Presence of Psb28 in wild-type membranes incubated in 25 mm Tris/HCl, pH 7.0, containing no addition (control), 1 m CaCl2, 0.1 m Na2CO3, 0.1 m NaOH, or trypsin (50 μg mL−1). The membranes were analyzed by SDS-PAGE and the gel was blotted to a PVDF membrane and D1 and Psb28 were detected by specific antibodies (blots). C, Membrane proteins from wild type were separated by 2D analysis, the gel was either stained (stained gel) or blotted to a PVDF membrane, and Psb28 was detected by specific antibodies (blot). Designation of complexes: PSI(3) and PSI(1), trimeric and monomeric PSI complexes, respectively; RCC(2) and RCC(1), dimeric and monomeric PSII core complexes, respectively; RC47, PSII core complex lacking CP43; U.P., Unassembled proteins.
Figure 2.
Identification of Psb28 in the PSII complexes of the phycobilisome-less strain PAL (A), the site-directed mutant D1-Asn-359His (B), and the psbC/slr0228 double mutant ΔCP43/ΔSlr0228 (C). Membrane proteins of the strains were separated by 2D BN/SDS-PAGE. PAL proteins in the gel were stained by Coomassie Blue, proteins of the D1-Asn-359His, and ΔCP43/ΔSlr0228 mutants were blotted onto PVDF and immunodetected by antibodies against CP47 and Psb28. The arrow indicates the stained Psb28 protein, which was identified by mass spectrometry in the PAL mutant. Six micrograms of Chl were loaded for each sample.
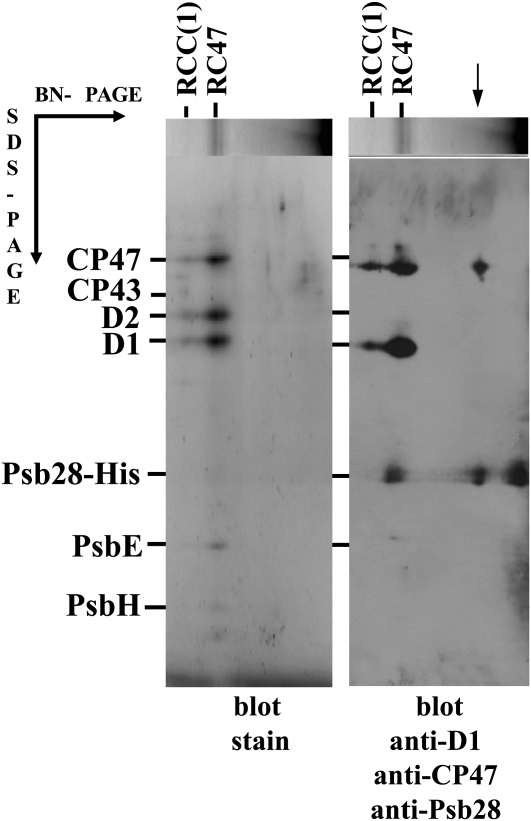
Further evidence for the preferential binding of Psb28 to RC47 was obtained using a strain in which the original psb28 gene was replaced by a His-tagged copy. Thylakoids from this Psb28-His/ΔPsb28 strain were solubilized with dodecyl maltoside (DM) and the extract was loaded on the nickel-affinity chromatography column. Bound proteins were analyzed by 2D BN/SDS-PAGE (Fig. 3A) and immunoblotting using antibodies against Psb28, D1, CP47, and CP43 (Fig. 3B). The majority of Psb28-His was present as free protein, but a significant amount was found in RC47. The isolated preparation also contained a small amount of RCC(1). Interestingly, a significant fraction of Psb28-His was associated with the unassembled CP47 confirming the previous indications obtained by analysis of the mutants D1-Asn-359His and ΔCP43/ΔSlr0228.
Figure 3.
Analysis of the PSII complexes isolated from the Psb28-His/ΔPsb28 strain. PSII complexes isolated by nickel-affinity chromatography from the Psb28-His/ΔPsb28 strain expressing the His-tagged Psb28 were analyzed by 2D BN/SDS-PAGE and proteins were blotted onto a PVDF membrane, stained (blot stain), and probed with antibodies against CP47, D1, and Psb28. The complex of CP47 and Psb28-His is designated by the arrow.
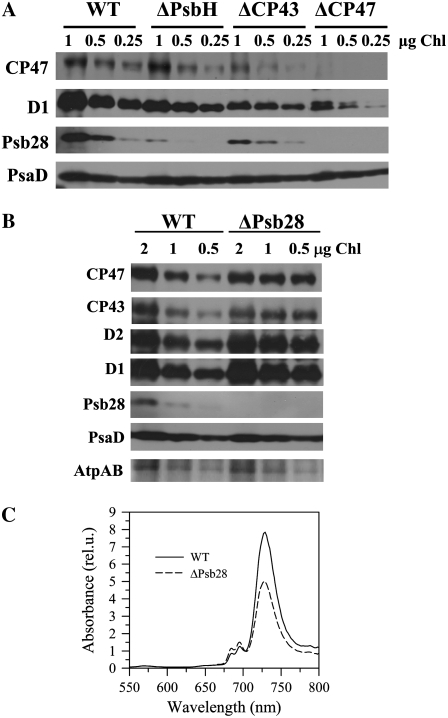
Investigation of the Psb28 content in several mutants differing in the content of PSII complexes again showed good correlation between Psb28 and CP47. In the mutant ΔCP43, which assembles only RC47 due to the deleted psbC gene, the decreased amount of Psb28 corresponded to about 20% to 30% of the wild-type level of CP47 (Fig. 4A). In the strain lacking CP47, the Psb28 became undetectable. Interestingly, in the absence of a small PSII subunit PsbH, which is bound to CP47, the level of Psb28 was also strongly decreased suggesting the requirement of PsbH for binding of Psb28 to CP47 (Fig. 4A).
Figure 4.
Accumulation of Psb28 in several PSII mutants differing in accumulation and stability of CP47 (A), level of PSII and PSI in cells of wild-type (WT) and ΔPsb28 analyzed either by western blotting (B) or by 77K Chl fluorescence emission spectra (C). A, Membranes from WT, psbH deletion mutant ΔPsbH, psbC deletion mutant ΔCP43, and psbB deletion mutant ΔCP47 were analyzed by denaturing SDS-PAGE and CP47, D1 and Psb28 were detected using specific antibodies. Correct protein loading was shown by immunodetection with PsaD-specific antibody. 1, 0.5, and 0.25 μg of Chl were loaded onto the gel for each sample. B, Membranes from WT and the psb28 deletion mutant ΔPsb28 grown in the presence of Glc were analyzed by denaturing SDS-PAGE and CP47, CP43, D2, D1, Psb28, and PsaD were detected by specific antibodies. Correct protein loading was shown by protein staining of α- and β-subunits of ATP synthase (AtpAB). 2, 1, and 0.5 μg of Chl were loaded onto the gel for each sample. C, 77K fluorescence spectra of WT cells (solid line) and ΔPsb28 (dashed line) cultivated in the presence of Glc were obtained using an Aminco Bowman Series 2 luminescence spectrometer (Spectronic Unicam). The same number of cells for each strain was excited at 435 nm, spectra were corrected for the sensitivity of the photomultiplier, and normalized to 570 nm (the fluorescence peak of rhodamine used as an internal standard).
The relationship between CP47 and Psb28 was further verified by constructing two double mutants in which the deletion of the psb28 gene was performed in strains lacking the psbC gene and the psbEFLJ operon. In the psbC deletion strain ΔCP43, additional inactivation of the psb28 gene led to a 50% decrease in the RC47 level and the unassembled CP47 became almost undetectable (Supplemental Fig. S1). Because the radioactive labeling of D1 was similar in both strains, we assume that the lower level of RC47 is caused by low availability of the newly synthesized CP47 in the absence of Psb28. Accordingly, in the psbEFLJ/psb28 double mutant ΔCYT/ΔPsb28, which accumulates the unassembled CP47, but no other CP47-containing PSII complexes (Komenda et al., 2004), the deletion of the psb28 gene led to a substantial decrease in the steady-state level of the unassembled CP47, whereas the accumulation of the unassembled D1 was increased (Supplemental Fig. S2).
Absence of Psb28 Leads to the Low Cellular Content of PSI But Does Not Affect the Function and Repair of PSII
To elucidate the role of Psb28 in the structure and function of PSII, we characterized the psb28 single deletion mutant (ΔPsb28) in which the psb28 gene was replaced with a zeocin-resistance cassette. The mutant exhibited slower photoautotrophic growth in comparison with wild type and contained a lower cellular level of Chl, especially under low light conditions. However, its light-saturated rate of the overall photosynthesis on a per cell basis was similar to that in wild type (Table I) and the rates of QA reoxidation measured in the presence and absence of the PSII inhibitor diuron, as well as thermoluminescence characteristics, were identical in wild type and ΔPsb28 (data not shown). These results showed that Psb28 is not important for the function of the fully assembled photochemically active PSII complexes and it corresponded well with the absence of the protein in the dimeric PSII core complex. On the other hand, the rate of PSII-mediated oxygen evolution in the mutant cells largely exceeded that in wild-type cells and this rate was even higher when measured on the per Chl basis, suggesting a decreased level of PSI in the mutant cells (Table I). This was confirmed by a semiquantitative western blot (Fig. 4B), which showed 20% to 40% higher PSII protein levels in the mutant (shown for all four large PSII proteins D1, D2, CP43, and CP47) when the gel was loaded on the basis of Chl content. Because about 80% of Chl in Synechocystis is bound in PSI, the blot did not show a significant decrease in the level of the PSI protein PsaD in the mutant. The results of the protein analyses were supported by the 77K Chl fluorescence emission spectra of cells from both strains (Fig. 4C). The spectra were taken using the same optical density of cells and were normalized using rhodamine as the internal standard (Sobotka et al., 2008). The significantly lower fluorescence maximum of PSI at 725 nm in mutant cells unequivocally confirmed the lower cellular level of PSI.
Table I.
Characteristics of the Synechocystis 6803 wild type and the psb28 deletion mutant ΔPsb28 grown at irradiance 10 (LL) or 40 (ML) μmol photons m−2 s−1
| Strains | Autotrophic Doubling Timea | Cellular Chl Contentb | Photosynthetic Oxygen Evolution (OD)c | PSII Oxygen Evolution (OD)d | PSII Oxygen Evolution (Chl)e |
|---|---|---|---|---|---|
| Wild type LL | 18.3 ± 0.1 | 2.59 ± 0.05 | 993 ± 151 | 588 ± 89 | 216 ± 35 |
| Wild type ML | 8.3 ± 0.1 | 2.54 ± 0.07 | 1,150 ± 191 | 935 ± 55 | 374 ± 21 |
| ΔPsb28 LL | 22.8 ± 0.2 | 1.60 ± 0.06 | 945 ± 141 | 1,283 ± 59 | 830 ± 54 |
| ΔPsb28 ML | 11.4 ± 0.1 | 1.87 ± 0.04 | 1,020 ± 185 | 1,353 ± 141 | 719 ± 72 |
Doubling time in hours measured in microtitration plates; means of 10 measurements ± sd; initial OD750 nm of the cultures was 0.005.
Chl content in micrograms per OD750 in the cells of both strains cultivated in the presence of Glc; means of three measurements ± sd.
Light-saturated rate of oxygen evolution in nanomoles O2 per OD750 nm and hours in cells of both strains cultivated in the presence of Glc; means of three measurements ± sd.
Light-saturated rate of oxygen evolution in nanomoles O2 per OD750 nm and hours in cells of both strains cultivated in the presence of Glc and measured in the presence of 1 mm p-benzoquinone and 5 mm potassium ferricyanide; means of three measurements ± sd.
Light-saturated rate of oxygen evolution in micromoles O2 per mg Chl and hours in cells of both strains cultivated in the presence of Glc and measured in the presence of 1 mm p-benzoquinone and 5 mm potassium ferricyanide; means of three measurements ± sd.
Because Psb28 was recognized as a component of RC47 in which the selective replacement of D1 most probably takes place (Komenda et al., 2004, 2006), we were interested in whether the absence of Psb28 affects D1 turnover. Pulse-chase experiments (Fig. 5) showed that the turnover of D1 was slightly accelerated in the absence of Psb28. Pulse-chase labeling revealed a half-life of D1 in wild type of about 2.5 h, whereas in the mutant ΔPsb28 it was approximately 2 h. To find out whether the resistance of PSII photochemistry to photodamage was also similar in both strains, we evaluated the time course of light-induced inhibition of oxygen evolution in wild-type and mutant cultures subjected to white light of 500 μmol photons m−2 s−1, either in the absence or presence of the protein synthesis inhibitor lincomycin. No differences in the decline of oxygen evolution were observed between wild type and ΔPsb28 during both measurements (Fig. 6A). Recovery from photoinhibition under low light conditions was even faster in ΔPsb28 than in wild type as proven by assessment of oxygen evolution in photoinhibited cells of wild type and ΔPsb28 during subsequent incubation at 50 μmol photons m−2 s−1 (Fig. 6B). The results showed that the oxygen-evolving PSII complexes of the mutant are equally sensitive to light-induced inactivation as the complexes in wild type, whereas the repair of PSII seems to be slightly more efficient, corresponding with the accelerated turnover of D1 observed in the strain.
Figure 5.
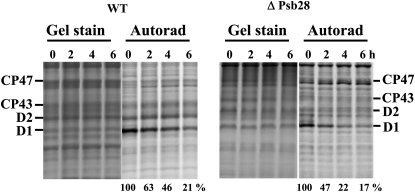
Degradation of the PSII proteins in wild-type (WT) and ΔPsb28 strains under high irradiance monitored by radioactive pulse-chase labeling. Autotrophic cells of both strains were subjected to 250 μmol photons m−2 s−1 of white light for 20 min in the presence of [35S]Met/Cys. The cells were then washed, supplemented with cold Met/Cys, and subjected to 500 μmol photons m−2 s−1 of white light for 6 h. Thylakoids were isolated and run on an SDS-PAGE gel. The gel was stained (gel stain) and radioactive labeling of the proteins was visualized using a phosphor imager (Autorad). Quantification of radioactivity in the D1 band was performed using ImageQuant software with samples of each strain equally loaded on Chl basis (2 μg of Chl; see gel stain) in a single gel. The radioactivity incorporated into the D1 band of each strain during pulse was taken as 100%; numbers show the means of the three measurements, sd did not exceed 6%.
Figure 6.
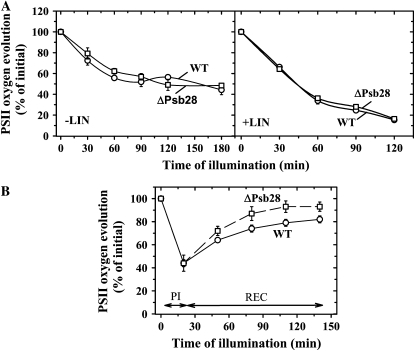
PSII repair under high and low irradiance in cells of wild type (WT) and ΔPsb28. A, Autotrophic WT cells (circles) and ΔPsb28 (squares) were illuminated with 500 μmol photons m−2 s−1 of white light for 180 min without antibiotic (left) or for 120 min in the presence of 100 μg mL−1 lincomycin (LIN; right). During illumination, PSII oxygen-evolving activity was assayed in whole cells. B, WT (circles) and ΔPsb28 (squares) cells were illuminated at 2,000 μmol photons m−2 s−1 for 20 min (PI), then the cells were transferred to low irradiance of 50 μmol photons m−2 s−1 and incubated for additional 120 min (REC). During these light regimes PSII oxygen-evolving activity was assayed in whole cells. Values in the plots represent the mean of three independent measurements ± sd. Initial values for WT and ΔPsb28 were in the range shown in Table I.
We were also interested in whether the absence of Psb28 affects levels of PSII proteins under extreme light conditions (2,000 μmol photons m−2 s−1). In wild-type cells, these conditions did not lead to significant changes in the content of PSII proteins, whereas in the ΔPsb28 strain a decline of about 50% of the level of D1 was observed (Supplemental Fig. S3A). We also compared the rate of PSII degradation in wild type and ΔPsb28 under conditions where protein synthesis was stopped by lincomycin (Supplemental Fig. S3B). No apparent differences were observed between both strains in the rate of degradation of D1, D2, CP43, CP47, and the α-subunit of cyt b-559. In the wild-type strain, the Psb28 protein was also degraded, whereas the PSI protein PsaD was stable again, confirming the relationship between Psb28 and PSII.
Psb28 Null Mutant Exhibits Limitation in the Synthesis of Chl and Chl-Binding Proteins CP47 and PsaA/PsaB Heterodimer
To characterize PSII assembly in wild type and ΔPsb28, we pulse labeled the cells with [35S]Met/Cys and analyzed their membrane protein complexes and incorporation of the radioactive label into the protein subunits by 2D BN/SDS-PAGE. In both strains, >90% of the D1 subunit was present in the dimeric and monomeric core complexes, RCC(2) and RCC(1) (Fig. 7). In wild type, at least 10% of D1 was present in the RC47, whereas in ΔPsb28 this complex was almost absent. In wild type, about 5% of overall CP47 was found in the fraction of unassembled proteins (Fig. 7, arrow 1), whereas no free CP47 was found in ΔPsb28. In agreement with the semiquantitative western blot, the overall amount of the stained PSII proteins in the gel loaded on the basis of Chl content was higher in the mutant than in wild type.
Figure 7.
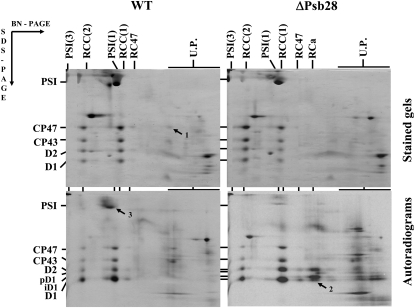
2D analysis of membrane protein complexes of wild type (WT) and the psb28 deletion strain ΔPsb28. Membranes isolated from radioactively labeled cells of WT and ΔPsb28 grown in the presence of Glc were analyzed by 2D electrophoresis. The obtained gels were stained and exposed to a phosphor imager plate. Complexes are designated as in Figure 1. RCa is a reaction center complex containing D1 and D2, but no CP43 and CP47 inner antennae. pD1 and iD1 indicate the unprocessed and partially processed forms of D1, respectively. Arrows designate important differences between the strains in the accumulation of CP47 (1) and RCa (2); and in the labeling of PSI subunits PsaA/PsaB (3). Each loaded sample contained 6 μg of Chl.
Autoradiograms obtained from the same 2D gels showed that, in wild type, the D1 protein was preferentially labeled in both RCC(2) and RCC(1). There was also apparent labeling of D1 in RC47, but not in the RCa complex containing D1, D2, cyt b-559, and PsbI (Fig. 7, autorad; see also Dobáková et al., 2007). In contrast, in membranes of ΔPsb28 about 20% of the labeled D1 was detected in the RCa complex and 30% in the fraction of unassembled proteins (Fig. 7, autorad, arrow 2). Our previous analyses showed that the presence of the RCa complex and unassembled D1 protein in various Synechocystis mutants indicates that the PSII assembly is limited by availability of CP47 (Komenda et al., 2004, 2005; Dobáková et al., 2007). In agreement with this interpretation, the labeled unassembled CP47 was present in wild type, but absent in ΔPsb28. Surprisingly, the absence of Psb28 also negatively affected the synthesis of a pair of large Chl-binding subunits, the PsaA/PsaB heterodimer of PSI. The radioactive labeling of this heterodimer in wild type exceeded that in ΔPsb28 by several times (Fig. 7, autorad, arrow 3). Overall, the results of radioactive labeling suggested that Psb28 is needed for the efficient synthesis of Chl proteins CP47 and PsaA/PsaB.
To explain the lower Chl level and most probably the related limitation in synthesis of CP47 and PsaA/PsaB observed in the ΔPsb28 mutant, precursor/degradation products of the Chl biosynthetic pathway were quantified in cells of each strain. As shown in Table II, mutant cells contained about 20-fold higher level of protoporphyrin IX than wild type and a substantial amount of this pigment was released by the ΔPsb28 cells into the growth medium (approximately 0.58 mg of protoporphyrin IX in 1 L of cell culture at approximately 0.5 OD750 in comparison with 0.057 found in wild-type culture). Level of magnesium (Mg) protoporphyrin IX monomethyl ester was also highly increased in the mutant cells; in contrast, accumulation of later Chl precursors, protochlorophyllide and chlorophyllide, reached only about 60% to 70% of the wild-type levels. This lower accumulation corresponds well with the total Chl level in the mutant, indicating that the insufficient synthesis of protochlorophyllide limits Chl availability in the ΔPsb28 strain. Consistently, the level of pheophorbide, a product of Chl catabolism, was found to be similar in the mutant and the wild type, suggesting that Chl degradation is not enhanced by deletion of the psb28 gene (Table II). The data suggested that, in the absence of Psb28, synthesis of Chl is significantly less efficient and this inefficiency could be directly related to the inhibition of synthesis of CP47 and PsaA/PsaB. Interestingly, synthesis of the other Chl proteins CP43 and D2 was not affected by the absence of Psb28 and synthesis of D1 was even more pronounced (see Fig. 7; Supplemental Fig. S2).
Table II.
Analysis of components of the Chl biosynthesis/degradation pathway in the wild-type strain and the ΔPsb28 mutant
Values represent mean ± sd of at least three independent experiments using different cultures harvested in logarithmic growth phase.
| Strain | Wild Type | ΔPsb28 |
|---|---|---|
| % of wild type | ||
| Protoporphyrin IX | 100 ± 18 | 2,340 ± 340 |
| Mg-protoporphyrin IX | 100 ± 12 | 157 ± 19 |
| Mg-protoporphyrin IX monomethyl ester | 100 ± 21 | 670 ± 82 |
| Protochlorophyllide | 100 ± 13 | 64 ± 8 |
| Chlorophyllide | 100 ± 32 | 68 ± 12 |
| Chl | 100 ± 9 | 72 ± 6 |
| Pheophorbide | 100 ± 22 | 119 ± 9 |
DISCUSSION
Role of Psb28 in the Structure and Function of PSII
The Psb28 protein has been previously detected as a nonstoichiometric component of the PSII preparation purified using nickel-affinity chromatography from a Synechocystis strain expressing His-tagged CP47 (Kashino et al., 2002). Please note that this purification method pulls down all PSII complexes containing CP47-His, including RCC(2), RCC(1), RC47, and unassembled CP47 and therefore it is not clear to what PSII complex Psb28 actually binds. Very recently, Sakurai et al. (2007) have also detected the protein in the isolated monomeric PSII complex. However, this complex was obtained by glycerol density gradient centrifugation, which in our hands has insufficient separation ability to resolve RCC(1) and RC47; therefore, the putative monomeric PSII most probably contained both complexes. Here, we show that Psb28 binds preferentially to RC47 and only the very small amount of Psb28 was occasionally found in the monomeric RCC(1) complex. The presence of Psb28 in RCC(1) was typical for strains containing a large amount of RC47, indicating that the protein most probably remains attached for a limited time after binding of CP43 to RC47. However, we have never found the protein in the dimeric complex RCC(2), which is considered to be the native, fully functional competent form of PSII (Barber, 2006). In accordance with this localization, the functional properties of PSII in the psb28 deletion mutant were not negatively affected by the absence of the protein. On the contrary, the mutant PSII activity on a per cell basis even exceeded that in the wild type. We believe that this high PSII activity is related to the very effective PSII repair cycle that operates in the mutant. This cycle is even more efficient than in wild type as documented by the accelerated turnover of the D1 protein and by the faster low light-induced recovery of PSII activity after photoinhibition of the mutant cells (Figs. 5 and 6). The fast repair cycle maintains all PSII complexes of the mutant in the perfectly functional state, whereas in wild type, some PSII complexes might be waiting for repair and therefore do not contribute to the overall activity.
The strain expressing His-tagged Psb28 allowed us to preferentially isolate the RC47 complex. However, the eluate from the chromatographic column also contained a complex of Psb28-His with unassembled CP47. Because no RC complexes or unassembled D1 or D2 was detected in the preparation, this complex could not originate from disassembly of RC47 and we assume that it represents a native complex present in vivo. On the other hand, on 2D gels we never observed the complex of CP47 with the nontagged Psb28. However, in the strains with high levels of unassembled CP47 like D1-Asn-359His, the unassembled Psb28 protein is found as a diffuse band smearing from the gel region approximately corresponding to the mobility of the unassembled CP47, suggesting that the CP47-Psb28 complex exists, but is unstable and falls apart during native electrophoresis. We propose that CP47 and Psb28 form a rather unstable precomplex, which is stabilized by the presence of the His-tag attached to Psb28. In this respect, it is interesting that the level of Psb28 is significantly decreased in the mutant lacking the PsbH protein (Fig. 3A), which was previously shown to be essential for the binding of Hli proteins to CP47 (Promnares et al., 2006). We therefore assume that the Psb28 binding site on CP47 is located in the vicinity of PsbH. Unfortunately, we were not able to verify this proposal using nitrilotriacetic acid-gold labeling of Psb28-His because the protein is present mostly in RC47, which is too small for successful single particle analysis using electron microscopy.
Role of Psb28 Protein in the Synthesis of Chl and Chl-Binding Proteins
The inhibition of the CP47 and PsaA/PsaB synthesis in the psb28 deletion mutant is paralleled by the perturbation in the Chl biosynthesis manifested by the accumulation of intermediates preceding protochlorophyllide formation. There are two possible explanations for this observation: (1) Psb28 is required for efficient synthesis of CP47 apoprotein or for Chl binding to it and in its absence the newly synthesized Chl is not consumed and its biosynthesis is stopped at the stage of formation of the isocyclic ring E; (2) Psb28 is needed for the final stage of biosynthesis of Chl that is utilized for synthesis of CP47. The first possibility assuming the direct action of Psb28 on the CP47 apoprotein synthesis seems to be improbable because it assumes the regulatory effect of the unused Chl at one of the latest stages of the biosynthesis pathway. Biosynthesis of heme is known to be regulated at early stages by the end product of the pathway (i.e. heme) in order to prevent accumulation of potentially toxic intermediates and similar mechanism seems to regulate the synthesis of Chl in plants (Meskauskiene et al., 2001). As Mg protoporphyrin monomethyl ester is converted to protochlorophyllide by a poorly understood aerobic cyclase enzyme (Minamizaki et al., 2008), the second explanation would presume a direct or indirect role of Psb28 in the cyclase reaction. In this respect, it is interesting that we have detected a substantial amount of the putative aerobic cyclase encoded by the sll1214 gene (Minamizaki et al., 2008), as well as protochlorophyllide oxidoreductase in the purified PSII core complex (data not shown). We therefore hypothesize that the Psb28 protein assists the efficient conversion of Mg protoporphyrin monomethyl ester into Chl, which is immediately bound to the CP47 apoprotein.
Although it has been clearly documented that the Psb28 protein is associated with PSII, the absence of Psb28 was also manifested by the limited synthesis of the PsaA/PsaB heterodimer of PSI and by the consequent decrease in the cellular content of PSI. This result suggested the existence of a link between the synthesis of CP47 and PsaA/PsaB. On the other hand, the absence of Psb28 did not affect the synthesis of D2 and CP43 and paradoxically resulted in the acceleration of synthesis and degradation of another PSII Chl-binding protein D1 as documented by the fast PSII repair (Figs. 5 and 6) and by the radioactive protein labeling (Fig. 7). Recent results showing the association of so-called high light-induced proteins with PSII (Promnares et al., 2006; Yao et al., 2007) and their presumable role in the temporary binding of Chl during the PSII repair-related D1 replacement (Vavilin et al., 2007) indicate that Chl molecules released from the degraded D1 can be reused. It is possible that the lack of the newly synthesized CP47 allows more Chl precursors to be utilized for the synthesis of Chl consumed during selective D1 turnover. Binding of the newly synthesized Chl would suppress Chl reutilization as an expected rate-limiting step of the repair process. Consequently, the PSII repair becomes faster in the absence of Psb28.
The differences among various Chl-binding proteins in the response to the partial inhibition of Chl synthesis could be explained by their different affinity for Chl. We have recently reported the selective inhibition of the CP47 synthesis in comparison to other PSII subunits in a Synechocystis Chl-deficient mutant lacking the Gun4 protein required for Mg chelatase activity (Sobotka et al., 2008). However, this mutant contains only about 20% of the wild-type level of Chl, whereas the Psb28-less strain contains at least 60% of the wild-type level, but still the synthesis of CP47 is selectively (at least as concerns PSII proteins) inhibited. The different interpretation of these results could be based on the branching and spatial separation of the Chl biosynthesis pathway at the final stages of the process, most probably at the formation of the Chl-specific E ring. In this way, only the branch terminating at CP47 and possibly PsaA/PsaB could be affected in the ΔPsb28 strain, whereas the other branch ending at D2 and CP43 remains unaffected or, as in the case of the D1 protein, is even accelerated due to the increased availability of the cyclase substrate. The data from Arabidopsis indicating the localization of the Chl cyclase in both the envelope membrane as well as in thylakoids support this view (Tottey et al., 2003).
MATERIALS AND METHODS
Construction and Cultivation of Cyanobacterial Strains
The strains used in the study were derived from the Glc-tolerant strain of Synechocystis 6803 (Williams, 1988) referred to as wild type. The following, previously described strains were used in the study: (1) the CP43-less strain ΔCP43 (Vermaas et al., 1988); (2) the CP47 deletion strain ΔCP47 with the psbB gene replaced with the kanamycin resistance cassette; (3) the ΔCYT strain with the psbEFLJ operon replaced with a kanamycin resistance cassette (Pakrasi et al., 1988); (4) the psbH deletion mutant ΔPsbH with the psbH gene replaced with a kanamycin resistance cassette (Komenda and Barber, 1995); (5) phycobiliprotein-free strain PAL (Ajlani and Vernotte, 1998); (6) the site-directed mutant D1-Asn-359His (Kuviková et al., 2005); and (7) the psbC/slr0228 double deletion mutant ΔCP43/ΔSlr0228 (Komenda et al., 2006).
The ΔPsb28 strain was prepared by the replacement of most of the psb28 gene (nucleotides 18–298) with a zeocin resistance cassette using a megaprimer PCR method (Burke and Barik, 2003). We have adapted this method to generate linear deletion constructs containing upstream and downstream regions of the psb28 gene with the zeocin resistance cassette in the middle. In the first step, upstream and downstream regions of psb28 were separately amplified using long fusion primers complementary to the psb28 gene in one direction and the zeocin cassette in the other: (1) 5′-ACATTAATTGCGTTGCGCTCACTGCAAATTGAATTTCAGCCATG-3′ and (2) 5′-CAACTTAATCGCCTTGCAGCACATCGGCGCAGAAAATGGCTTAGG-3′ (the psb28 part is underlined). These fusion primers were used in pairs with psb28 forward and reverse primers: (1) 5′-TGTTCTACCTGCTCGATCGC-3′ and (2) 5′-GAGTTTCGCTACTATCAGCG-3′. In the second step, the zeocin resistance cassette (Streptoalloteichus hindustanus; Invitrogen) was amplified using PCR products from the first step as primers. Finally, the complete deletion construct was amplified using psb28 forward and reverse primers and used for transformation of Synechocystis 6803 cells. Transformants were selected and segregated on zeocin-containing agar plates; their full segregation was confirmed by PCR.
Multiple psb28 deletion strains were obtained by transformation of single mutants lacking psbC and psbEFLJ genes using chromosomal DNA from ΔPsb28 and their selection was based on additional resistance to zeocin.
The Psb28-His strain expressing Psb28 tagged with 6×His at its N terminus under control of the psbA2 promoter (Psb28-His/ΔPsb28) was constructed using the pSBA plasmid (Lagarde et al., 2000) in a procedure analogous to that described by Tichý et al. (2003). The resulting strain synthesizing both wild-type and His-tagged forms of Psb28 was transformed with chromosomal DNA from the psb28 deletion mutant and selected by resistance to zeocin. The complete deletion of the psb28 wild-type copies in the Psb28-His strain was confirmed by PCR.
Liquid cultures were grown in 100 to 200 mL of BG11 containing 5 mm Glc using 500-mL Erlenmeyer flasks, aerated using an orbital shaker, irradiated with 30 μmol photons m−2 s−1 of white light at 29°C, and used when they reached a Chl concentration of about 5 μg mL−1. Solid medium contained in addition 10 mm Tes/NaOH, pH 8.2, 1.5% agar, and 0.3% sodium thiosulfate (Pakrasi et al., 1988).
Photoinhibition experiments were performed with cells cultivated in double-wall, thermoregulated cultivation cylinders (internal diameter of 35 mm). Here, the culture was maintained at a Chl concentration of 6 to 8 μg mL−1 by regularly diluting with approximately 10 mL of BG11 medium every 150 min. The culture was bubbled with air containing 2% (v/v) CO2 and illuminated with white light at 40 μmol photons m−2 s−1 at 29°C. For the large-scale cultivation used for isolation of PSII complexes, cultures were grown in 10-L flasks (culture volume 6–8 L), stirred by a magnetic stirrer, and bubbled with air.
Measurements of autotrophic growth rates were performed in microtitration plates (culture volume 0.25 mL) under intensive shaking. Optical densities at 750 nm were measured every 6 h using a microplate reader (Tecan Sunrise). Values plotted against time were used for calculation of the doubling time.
Fluorometric and Polarographic Methods
The FV/FM parameter and kinetics of Chl variable fluorescence decay were measured in dark-adapted cultures (2.5 μg Chl mL−1) using a modulation PAM101 fluorometer (Walz) with an ED-101US cuvette and the dual-modulation kinetic fluorometer (Photon Systems Instruments), respectively (Tichý et al., 2003). The light-saturated steady-state rate of oxygen evolution in cell suspensions was measured polarographically in BG11 medium containing 10 mm HEPES/NaOH, pH 7.0, using 0.5 mm p-benzoquinone and 1 mm potassium ferricyanide as artificial electron acceptors.
Preparation of Membranes and Their Protein Analyses
Cyanobacterial membranes were prepared by breaking the cells using glass beads (Komenda and Barber, 1995) with the following modifications: Cells were washed, broken, and finally resuspended in 25 mm MES/NaOH, pH 6.5, containing 10 mm CaCl2, 10 mm MgCl2, and 25% glycerol. The large-scale isolation of PSII complexes for chromatographic purification was performed using a similar procedure in which the cells were resuspended in 20 mL of thylakoid buffer (25 mm MES, pH 6.5, 100 mm NaCl) containing the protease inhibitor cocktail (Roche), the same volume of glass beads was added and the cells were broken eight times for 15 s in the smallest bead-beater container (Biospec Products) with 5-min interruption for cooling on ice. Glass beads were subsequently removed by filtering and thylakoids were obtained by differential centrifugation.
For separation of membrane and soluble fractions, the broken cells were pelleted (20,000g, 15 min) and the sediment (membrane fraction) was resuspended in the original volume of 25 mm MES/NaOH, pH 6.5, containing 10 mm CaCl2, 10 mm MgCl2, and 25% glycerol. The supernatant represented the soluble fraction.
To distinguish membrane-embedded and membrane-associated proteins, the membranes (5 μg Chl) were spun down and resuspended in 25 mm Tris/HCl, pH 7.0, containing 1 m CaCl2, in 0.1 m Na2CO3, or in 0.1 m NaOH. The suspensions were left on ice with occasional mixing for 4 h, then spun down, resuspended in 25 mm Tris/HCl, pH 7.0, containing 1 m Suc, and analyzed by SDS-PAGE. The tryptic digestion of thylakoids was performed as described in Komenda et al. (2002). After incubation for 4 h, the protease was inhibited with 2 mm Pefabloc SC.
The protein complexes isolated from the thylakoid membranes were solubilized with DM (DM/Chl = 40 [w/w]) and analyzed by BN electrophoresis at 4°C in a 5% to 14% gradient polyacrylamide gel according to Schägger and von Jagow (1991). Samples with the same Chl content (6 μg for gel staining and 1 μg for western blot) were loaded onto the gel.
The protein composition of the complexes was analyzed by electrophoresis in a denaturing 12% to 20% linear gradient polyacrylamide gel containing 7 m urea (Komenda et al., 2002). Complete lanes from the native gel were excised, incubated for 30 min in 25 mm Tris/HCl, pH 7.5, containing 1% SDS (w/v), and placed on top of the denaturing gel; two lanes were analyzed in a single denaturing gel. Proteins separated in the gel were either stained by Coomassie Blue or transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were incubated with specific primary antibodies and then with secondary antibody-horseradish peroxidase conjugate (Sigma). The primary antibodies used in the study were raised in rabbits against (1) residues 58 to 86 of the spinach (Spinacia oleracea) D1 polypeptide; (2) the last 12 residues of the D2 polypeptide from Synechocystis 6803; (3) residues 380 to 394 of barley (Hordeum vulgare) CP47; (4) whole isolated CP43 from Synechocystis 6803; and (5) the last 15 C-terminal residues of the Psb28 protein from Synechocystis 6803. For autoradiography, the gel or the membrane with labeled proteins was exposed to x-ray film at laboratory temperature for 2 to 3 d or to a phosphor imager plate (GE Healthcare) overnight.
Samples with the same Chl content were used for the direct comparison and quantification of stained or labeled proteins and were run on a single gel. Quantification of bands was performed using ImageQuant 5.2 software (GE Healthcare).
Radioactive Labeling of Cells
For radioactive labeling, cells containing 75 μg of Chl were resuspended and shaken in 250 μL of BG11 in a microcentrifuge tube for 30 min under illumination with 60 μmol photons m−2 s−1 and then a mixture of [35S]Met and [35S]Cys (Trans-label; MP Biochemicals) was added (final specific activity 400 μCi mL−1). The suspension was exposed to light at irradiance and temperature indicated and afterward the cells were frozen in liquid nitrogen and used for isolation of thylakoids.
Isolation of Complexes Containing Psb28-His
PSII complexes containing Psb28-His were isolated from thylakoids of the Psb28-His/ΔPsb28 strain using affinity chromatography on immobilized Ni2+ ions. Thylakoid membranes were solubilized with DM, final concentration 2% (w/v) in thylakoid buffer (25 mm MES, pH 6.5, 100 mm NaCl) for 30 min in the dark and on ice. The Chl concentration was 1 mg mL−1. The unsolubilized material was removed by centrifugation for 20 min at 60,000g at 4°C and the supernatant was loaded onto a chromatography column with Fractogel EMD Chelate (Merck) charged with Ni2+ and equilibrated with thylakoid buffer containing 0.04% DM (w/v; Bumba et al., 2005). The column was subsequently washed with the same buffer until the eluate remained colorless and the fraction containing Psb28-His was eluted using the isolation buffer with added 20 mm imidazole and 0.04% DM.
Chl Content
For the measurement of Chl concentration, sedimented cells or membranes were extracted with 100% methanol and Chl content in the extract was calculated from the absorbance at 666 and 720 nm (Wellburn and Lichtenthaler, 1984).
Quantification of Chl Intermediates
For quantitative determination of Chl precursors in the cells, 100 mL of culture at OD750 = ∼0.5 was filtered through a 4-μm cellulose filter to remove all precipitated pigments in growth medium and harvested. Pigments were extracted from cell pellets by three successive extractions with methanol containing 0.1% of NH4OH. Combined supernatants (approximately 1 mL in total) were filtered through a 0.22-μm filter and 100 μL were immediately subjected to HPLC analysis using a Vydac 201TP54 column (250 mm × 4.6 mm, C-18 reversed-phase silica gel). The column was eluted with a linear gradient from 0% of solvent A (methanol and 0.5 m ammonium acetate, 4:6 [v/v]) to 60% of solvent B (methanol and acetone, 6:4 [v/v]) over 4 min followed by a linear gradient of solvent B. 100% of solvent B was reached after 28 min at a flow rate of 1.1 mL/min at 35°C. HPLC peaks corresponding to protoporphyrin IX, Mg-protoporphyrin IX, Mg-protoporphyrin IX monomethyl ester, protochlorophyllide, and chlorophyllide were identified from their absorption spectra and from comparison with authentic standards (Sigma Aldrich; Frontier Scientific). HPLC fractions containing Chl intermediates were collected and concentrations of the corresponding compounds determined fluorometrically using a Spectronic Unicam series 2 spectrofluorimeter. Pheophorbide was quantified spectroscopically.
Analysis of Pigments in Growth Medium
For identification and quantification of tetrapyrroles precipitated in growth medium, a 4-μm cellulose filter from the previous paragraph was frozen in liquid nitrogen, powdered in a 2-mL Eppendorf tube, and dissolved in 1 mL of alkaline acetone. Acetone was then extracted with 400 μL of hexane to remove Chl and presence of tetrapyrroles was analyzed spectroscopically and fluorometrically. Protoporphyrin IX was quantified spectroscopically using an authentic standard (Sigma).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. 2D BN/SDS-PAGE of radioactively labeled membrane proteins from psbC deletion mutant ΔCP43 and the double mutant ΔCP43/ΔPsb28.
Supplemental Figure S2. 2D BN/SDS-PAGE of radioactively labeled membrane proteins from psbEFLJ deletion mutant ΔCYT and the double mutant ΔCYT/ΔPsb28.
Supplemental Figure S3. Levels of PSII proteins in cells of wild type and ΔPsb28 exposed to high light in the absence and presence of lincomycin.
Supplementary Material
Acknowledgments
We are grateful to Prof. E.-M. Aro, Prof. L.A. Eichacker, and Prof. R. Barbato for donation of specific antisera, and Dr. E. Prachová, Mr. P. Zelík, and Dr. J. Knoppová for help with strain cultivations, with measurements of growth curves, and with 2D gels. We also thank Mr. Daniel Canniffe for reading the manuscript.
This work was supported by the Grant Agency of the Czech Republic (project no. 206/06/0322), by the Ministry of Education, Youth and Sports of the Czech Republic (project no. MSM6007665808), and by the Czech Academy of Sciences (grant nos. AV0Z50200510 and IAA400200801).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Josef Komenda (komenda@alga.cz).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Ajlani G, Vernotte C (1998) Construction and characterization of a phycobiliprotein-less mutant of Synechocystis sp. PCC 6803. Plant Mol Biol 37 577–580 [DOI] [PubMed] [Google Scholar]
- Barber J (2006) Photosystem II: an enzyme of global significance. Biochem Soc Trans 34 619–631 [DOI] [PubMed] [Google Scholar]
- Bumba L, Tichý M, Dobáková M, Komenda J, Vácha F (2005) Localization of the PsbH subunit in photosystem II from the Synechocystis 6803 using the His-tagged Ni-NTA nanogold labeling. J Struct Biol 152 28–35 [DOI] [PubMed] [Google Scholar]
- Burke E, Barik S (2003) Megaprimer PCR: application in mutagenesis and gene fusion. Methods Mol Biol 226 525–532 [DOI] [PubMed] [Google Scholar]
- Dobáková M, Tichý M, Komenda J (2007) Role of the PsbI protein in Photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 145 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TN, Maglaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303 1831–1838 [DOI] [PubMed] [Google Scholar]
- Irrgang KD, Shi LX, Funk Ch, Schröder WP (1995) A nuclear-encoded subunit of the photosystem II reaction center. J Biol Chem 270 17588–17593 [DOI] [PubMed] [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41 8004–8012 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barber J (1995) Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and is dependent on protein synthesis. Biochemistry 34 9625–9631 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, DeVries R, Mullineaux CW, Tichý M, Nixon PJ (2006) The FtsH protease slr0228 is important for quality control of Photosystem II in the thylakoid membrane of Synechocystis PCC 6803. J Biol Chem 281 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Lupínková L, Kopecký J (2002) Absence of the psbH gene product destabilizes the photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur J Biochem 269 610–619 [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Müller BC, Dobáková M, Granvogl B, Eichacker LA (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279 48620–48629 [DOI] [PubMed] [Google Scholar]
- Komenda J, Tichý M, Eichacker L (2005) The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into Photosystem II in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol 46 1477–1483 [DOI] [PubMed] [Google Scholar]
- Kuviková S, Tichý M, Komenda J (2005) A role of the C-terminal extension of the Photosystem II D1 protein in sensitivity of the cyanobacterium Synechocystis PCC 6803 to photoinhibition. Photochem Photobiol Sci 4 1044–1048 [DOI] [PubMed] [Google Scholar]
- Lagarde D, Beuf L, Vermaas W (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 66 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438 1040–1044 [DOI] [PubMed] [Google Scholar]
- Lorković ZJ, Schröder WP, Pakrasi HB, Irrgang KD, Herrmann RG, Oelmüller R (1995) Molecular characterization of PsbW, a nuclear-encoded component of the photosystem II reaction center complex in spinach. Proc Natl Acad Sci USA 92 8930–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, Op den Camp R, Apel K (2001) FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamizaki K, Mizoguchi T, Goto T, Tamiaki H, Fujita Y (2008) Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 283 2684–2692 [DOI] [PubMed] [Google Scholar]
- Nanba O, Satoh K (1987) Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc Natl Acad Sci USA 84 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakrasi HB, Williams JGK, Arntzen CJ (1988) Targeted mutagenesis of the psbE and psbF blocks photosynthetic electron transport: evidence for a functional role of cytochrome b-559 in photosystem II. EMBO J 7 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promnares K, Komenda J, Bumba L, Nebesarova J, Vacha F, Tichy M (2006) Cyanobacterial small chlorophyll binding protein ScpD (HliB) is located on the periphery of Photosystem II in the vicinity of PsbH and CP47 subunits. J Biol Chem 281 32705–32713 [DOI] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM (2005) Synthesis and assembly of thylakoid protein complex: multiple assembly steps of photosystem II. Biochem J 388 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I, Mizusawa N, Ohashi S, Kobayashi M, Wada H (2007) Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiol 144 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199 223–231 [DOI] [PubMed] [Google Scholar]
- Shi LX, Lorković ZJ, Oelmüller R, Schröder WP (2000) The low molecular mass PsbW protein is involved in the stabilization of the dimeric photosystem II complex in Arabidopsis thaliana. J Biol Chem 275 37945–37950 [DOI] [PubMed] [Google Scholar]
- Shi LX, Schröder WP (2004) The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim Biophys Acta 1608 75–96 [DOI] [PubMed] [Google Scholar]
- Sobotka R, Dühring U, Komenda J, Peter E, Gardian Z, Tichy M, Grimm B, Wilde A (2008) Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J Biol Chem 283 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichý M, Lupínková L, Sicora C, Vass I, Kuviková S, Prášil O, Komenda J (2003) Synechocystis 6803 mutants expressing distinct forms of the photosystem II D1 protein from Synechococcus 7942: Relationship between the psbA coding region and sensitivity to visible and UV-B radiation. Biochim Biophys Acta 1605 55–66 [DOI] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Schiller HV, Merchant S, Jensen PE (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavilin D, Yao D, Vermaas W (2007) Small cab-like proteins retard degradation of Photosystem II-associated chlorophyll in Synechocystis sp. PCC 6803. J Biol Chem 282 306–314 [DOI] [PubMed] [Google Scholar]
- Vermaas WFJ, Ikeuchi M, Inoue Y (1988) Protein composition of the photosystem II core complex in genetically engineered mutants of the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 17 97–113 [DOI] [PubMed] [Google Scholar]
- Wellburn AR, Lichtenthaler K (1984) Formulae and programme to determine total carotenoids and chlorophyll a and b of leaf extracts in different solvents. In C Sybesma, ed, Advances in Photosynthesis Research. Martinus Nijjhoff, Dordrecht, The Netherlands, pp 10–12
- Williams JGK (1988) Construction of specific mutations in PSII photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167 766–778 [Google Scholar]
- Yao D, Kieselbach T, Komenda J, Promnares K, Hernandez Prieto MA, Tichy M, Vermaas W, Funk C (2007) Localization of the small CAB-like proteins in Photosystem II. J Biol Chem 282 267–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.