Abstract
The anthers in flowers perform important functions in sexual reproduction. Several recent studies used microarrays to study anther transcriptomes to explore genes controlling anther development. To analyze the secretion and other functions of the tapetum, we produced transcriptomes of anthers of rice (Oryza sativa subsp. japonica) at six progressive developmental stages and pollen with sequencing-by-synthesis technology. The transcriptomes included at least 18,000 unique transcripts, about 25% of which had antisense transcripts. In silico anther-minus-pollen subtraction produced transcripts largely unique to the tapetum; these transcripts include all the reported tapetum-specific transcripts of orthologs in other species. The differential developmental profiles of the transcripts and their antisense transcripts signify extensive regulation of gene expression in the anther, especially the tapetum, during development. The transcriptomes were used to dissect two major cell/biochemical functions of the tapetum. First, we categorized and charted the developmental profiles of all transcripts encoding secretory proteins present in the cellular exterior; these transcripts represent about 12% and 30% of the those transcripts having more than 100 and 1,000 transcripts per million, respectively. Second, we successfully selected from hundreds of transcripts several transcripts encoding potential proteins for lipid exine synthesis during early anther development. These proteins include cytochrome P450, acyltransferases, and lipid transfer proteins in our hypothesized mechanism of exine synthesis in and export from the tapetum. Putative functioning of these proteins in exine formation is consistent with proteins and metabolites detected in the anther locule fluid obtained by micropipetting.
Anthers in flowers produce via meiosis haploid microspores, which mature to become pollen (Goldberg et al., 1993; Bewley et al., 2000). Each anther usually consists of four layers of cells enclosing a central locule (Goldberg et al., 1993; Scott et al., 2004) in which the microspores mature. Cells of the outer three layers are highly vacuolated and presumed to be metabolically less active. Cells of the innermost layer, the tapetum, have dense cytoplasm and are metabolically active. They regulate maturation of the microspores. Tapeta in most angiosperms, including all major crops, are categorized as secretory (Hesse et al., 1993). During early anther development, the tapetum secretes wall hydrolytic enzymes into the locule to hydrolyze the microspore tetrad wall, thereby releasing individual microspores. Throughout anther development, the tapetum supplies nutrients and likely other enzymes and nonenzyme proteins to nurture the microspores. In some species, the haploid pollen can be obtained in abundance for intense investigations, including delineation of the germination and elongation processes and analysis of the pollen transcriptomes (Cheung and Wu, 2001; Malik et al., 2007). In contrast, few studies have investigated the diploid anther cells, especially the mechanism of function of the tapetum.
Nevertheless, genes controlling the development of various anther cell layers in Arabidopsis (Arabidopsis thaliana), maize (Zea mays), rice (Oryza sativa), and lotus (Lotus japonicus) have been examined with use of flower or anther transcriptomes obtained from DNA microarrays (Amagai et al., 2003; Wang et al., 2005; Ma et al., 2006, 2007; Masuko et al., 2006; Alves-Ferreira et al., 2007; Carlsson et al., 2007; Chen et al., 2007; Wijeratne et al., 2007; Kang et al., 2008). Some of these studies were made with comparisons between wild types and anther mutants. The mutants have knockout nuclear or mitochondrial (cytoplasmic male sterility) genes, and the knockouts lead to disrupted development of anther cell layers and pollen, thus resulting in male sterility. Results of these analyses provide insight into the functioning of the mutated genes and reveal new regulatory genes controlling anther development. The use of DNA microarrays to generate transcriptomes has technical limitations, which include low numbers of gene transcripts and minimal quantification. Also, obtaining the small anthers from Arabidopsis and many other species for microarray studies is difficult; thus, whole flowers or anthers of one or mixed developmental stages have been used.
Analyses of transcriptomes of plant and non-plant materials have revealed natural antisense transcripts (NATs; Lapidot and Pilpel, 2006; Ma et al., 2006; Nobuta et al., 2007). About 15% to 29% of the sense transcripts in mammals and 7% to 36% in plants have corresponding NATs. The functions of NATs are largely unknown. Studies of limited numbers of genes producing NATs, plus theoretical considerations, have indicated that NATs could modulate expression of their respective genes via transcriptional interference, RNA masking, different double-stranded RNA (dsRNA)-dependent mechanisms, and effect on DNA methylation. Information on the precise developmental changes of the sense and antisense transcripts allows researchers to explore the functions of NATs more precisely, as has occurred for several mammalian systems. Such information on flower anthers has previously been absent.
The tapetum is involved in a secretory process for the synthesis of exine, the outer wall of pollen (for review, see Blackmore et al., 2007). Exine is a complex lipid polymer that resists mild chemical treatments to release its monomers for structural analysis. Recently, analytical results of exine monomers released from controlled hydrolysis have suggested that exine is composed of two monomers, hydroxyl-fatty acids (OH-FAs) and isopropanoids, linked together by ester and ether bonds (Ahlers et al., 2003; Morant et al., 2007). The enzymes involved in OH-FA (Cabello-Hurtado et al., 1998; Kandel et al., 2006) and isopropanoid (Graham, 1998; Winkel-Shirley, 2001; Winkel 2004) synthesis in plants have been studied. However, the polymerization of OH-FA and isopropanoids and the overall mechanism of transfer of water-insoluble exine monomers, intermediates, and final products from the tapetum to the microspore surface are largely unknown.
We used the advanced sequencing-by-synthesis (SBS) technology to obtain transcriptomes of rice anthers of defined developmental stages and mature pollen. Our SBS-obtained transcriptomes offer several advantages over microarray data on anthers, because they: (1) include most transcripts; (2) are high quality; (3) have precise quantity data; (4) detail developmental changes; and (5) provide quantitative changes of the NATs in relation to their sense transcripts during development. We capitalized these advantages to dissect two major cell/biochemical functions of the tapetum. First, we delineated the global secretion in the tapetum and pinpointed abundant tapetum-specific secretory proteins whose functions now can be explored. Second, we successfully selected from hundreds of transcripts several transcripts encoding potential proteins for lipid exine synthesis and transport for future investigation. The functioning of some of these proteins is consistent with the identified secretory proteins and metabolites in the locule fluid we obtained with micropipetting. Here, we report the findings.
RESULTS AND DISCUSSION
Anthers of Six Progressive Developmental Stages and Mature Pollen Were Prepared for Making Transcriptomes
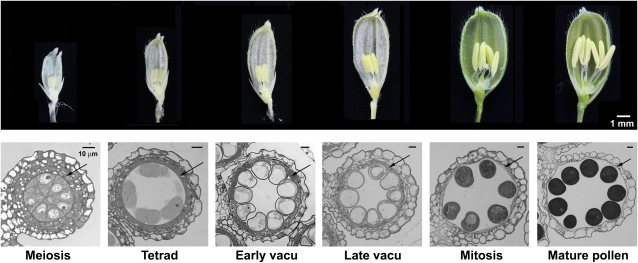
Transcriptome data of anthers and pollen are more reliable for analysis if detailed quantitative changes of every transcript in well-defined progressive developmental stages can be observed. In addition, the changes will provide information on the altered roles of the various genes in an anther during development and, in turn, the shifting functions of an anther and its components such as the tapetum and microspores. We obtained anthers of six developmental stages (Fig. 1), classified according to the cytological changes in microspores (Li et al., 2006), and mature pollen. RNA was extracted from these samples for preparation of SBS transcriptomes by Solexa.
Figure 1.
Photographs of spikelets and cross sections of anthers of six progressive developmental stages. Top row (dissecting microscopy), Spikelets with one side of the spikelet brackets removed to reveal the internal anthers. Bottom row (light microscopy), Cross sections of one lobe of an anther of each developmental stage, named according to the morphological features in the microspores. The arrow locates the tapetum layer enclosing the microspores in the locule. In the mature-pollen stage, the tapetum layer had disintegrated. In the bottom row, scale bar = 10 μm; its diminishing length in the images of the six progressive stages reflects the continuous enlargement of the anthers and microspores. vacu, Vacuolated.
High-Quality SBS Transcriptomes of Anthers and Mature Pollen Were Obtained
Earlier, transcriptomes of different rice organs were obtained with use of the Massive Parallel Signature Sequence (MPSS) technology (Nobuta et al., 2007). Only one of these organs, termed immature panicles, which are spikelets of developing flowers, is related to anthers. We used SBS, the sequencing technology advanced from MPSS, to construct transcriptomes of rice anthers and pollen.
In the seven SBS transcriptomes (Table I), the sequenced signatures (i.e. 20-mer tags of cDNA) vary from 1.1 to 4.3 million, which contain many duplicates. These sequenced signatures were filtered into 66,000 to 253,000 distinct signatures, which include nonoverlapping and overlapping sequences in the open reading frame (ORF), untranslated regions (UTRs), introns, and intergene spacers. The distinct signatures were assembled into specific transcripts of transcripts per million (TPM) values in each transcriptome (Table I). These genes number from 3 to 12 K in individual transcriptomes, for a total of 18,267 genes in all the transcriptomes. A small proportion of these genes were represented by NATs with no corresponding sense transcripts (to be described in a later section). According to TIGR annotation, the rice genome has 41,046 genes excluding transposable elements. Thus, the seven anther and pollen transcriptomes include transcripts of 44% (18,267/41,046) of the rice genes. This percentage should be higher in actuality, because we excluded transcripts with less than five TPM as possible background noises and also tags that hit identical genome sequences as mixed-pool noises (Bishop et al., 2005; Peiffer et al., 2008). Had we included transcripts having two to four TPM, we would have an additional 3,159 transcripts. Regardless, the percentage may be close to the theoretical maximal value for all anther transcripts, because other organs must have their respective organ-specific transcripts of genes (i.e. absent in anthers). Nevertheless, the anther transcriptomes exclude transcripts from genes that do not have the sequence GATC----, which is required for SBS sequencing. About 4.7% of the rice genes do not have the sequence GATC----.
Table I.
Parameters of the transcriptomes of anthers of six progressive developmental stages and pollen
Sequenced signatures are 20-mer sequences, which are filtered into distinct signatures and then distinct transcripts of specific genes. Transcripts are categorized according to their abundance in TPM. Anther transcripts that are >5× pollen transcripts in TPM represent largely those in sporophytic anther cells, and pollen transcripts that are >5× anther transcripts in TPM (average of those in the six anther stages) correspond to mainly those in gametophytic pollen. Anther and pollen transcripts that are >5× those in other rice organs in TPM represent highly anther/pollen-specific transcripts.
| Signatures or Transcripts | Developing Anthers
|
Pollen | |||||
|---|---|---|---|---|---|---|---|
| Meiosis | Tetrad | Early Vacuolated | Late Vacuolated | Mitosis | Mature Pollen | ||
| Total | |||||||
| Sequenced signatures | 4,307,036 | 1,774,208 | 1,726,668 | 1,598,476 | 1,150,116 | 1,128,456 | 3,805,478 |
| Distinct signatures | 253,065 | 182,491 | 196,274 | 145,672 | 125,415 | 117,430 | 66,324 |
| Distinct transcriptsa | |||||||
| TPM ≥5 | 12,635 | 6,916 | 9,245 | 6,664 | 4,584 | 3,895 | 3,175 |
| TPM ≥10 | 9,816 | 4,756 | 6,430 | 4,711 | 3,228 | 2,579 | 2,097 |
| TPM ≥100 | 1,385 | 717 | 880 | 798 | 533 | 466 | 544 |
| TPM ≥1,000 | 48 | 48 | 39 | 44 | 55 | 67 | 111 |
| Transcripts in anthers >5× in pollen, or in pollen >5× in (avg.) anthers | |||||||
| TPM ≥10 | 5,563 | 2,612 | 3,506 | 2,544 | 1,151 | 584 | 733 |
| TPM ≥100 | 828 | 488 | 585 | 524 | 169 | 63 | 333 |
| TPM ≥1,000 | 32 | 43 | 34 | 35 | 14 | 4 | 84 |
| Transcripts in anther-minus-pollen (from preceding rows) or pollen-minus-anther (from preceding rows) >5× in other organs | |||||||
| TPM ≥10 | 1,403 | 933 | 1,147 | 885 | 426 | 200 | 437 |
| TPM ≥100 | 247 | 248 | 288 | 262 | 89 | 31 | 247 |
| TPM ≥1,000 | 22 | 33 | 28 | 26 | 13 | 4 | 83 |
Both sense and antisense transcripts are included.
The quantities of transcripts in the various transcriptomes are normalized to TPM (Table I) so that we can compare the quantity of each transcript in different anther and pollen transcriptomes, as well as in MPSS transcriptomes of other rice organs (http://mpss.udel.edu). Table I subdivides the transcripts according to TPM abundance. Several dozens of the transcripts have TPM ≥1,000 (i.e. ≥0.1% total mRNA), and their encoded proteins would be needed in abundance to carry out functions.
TPM of Individual Transcripts Peaked in Level at Different Progressive Developmental Stages as Confirmed by RT-PCR
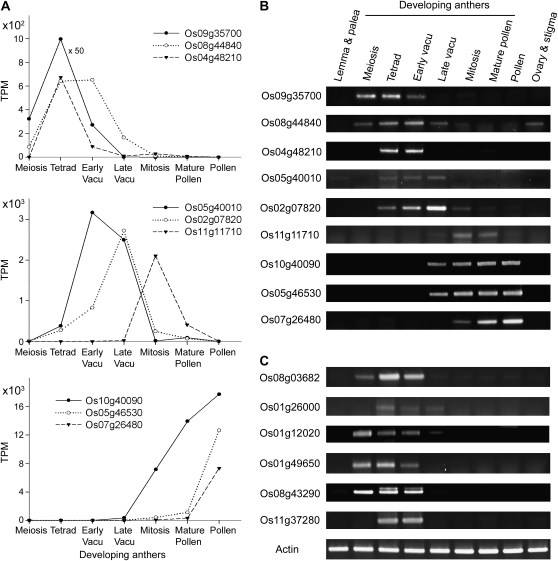
Figure 2A illustrates examples of transcripts with different patterns of developmental changes. Many of the transcripts whose levels increased at mid or late developmental stages had the highest levels in mature pollen. Most likely, these transcripts were present in microspores and continued to accumulate as the microspores matured to become pollen (Suen et al., 2003).
Figure 2.
Variations in the developmental profiles of anther transcripts. A, TPM of sample transcripts. B, RT-PCR results of the transcripts shown in A. C, RT-PCR results of special transcripts related to exine formation (Table V). Actin transcript was used as a loading control. The x axis of the developmental stages in this and other figures is not a linear time scale. Vacu, Vacuolated.
RT-PCR analyses of the above transcript examples confirmed the quantification via the transcript TPM (Fig. 2B). The pattern of expression increase, peaking, and decrease of each individual transcript faithfully matches the pattern plotted according to TPM.
The Anther Transcriptomes Possess NATs of Approximately 25% the Number of Sense Transcripts, and Individual Sense-Antisense Pairs May or May Not Share the Same Developmental Profiles
The rice anther transcriptomes include NATs corresponding to 12% to 25% of the sense transcripts (Table II). In individual transcriptomes, this percentage decreases with the more abundant transcripts, from TPM ≥5, 10, 100, to 1,000 (Table II). One possible explanation is that the less-abundant transcripts can be pivotal for developmental changes or enzymatic reactions and are easier to regulate with minimal amounts of NATs. For the abundant transcripts (TPM ≥10, 100, and 1,000), the proportion of sense transcripts with NATs increases from early to late development, possibly reflecting that many of these abundant transcripts were present first in microspores and then pollen. The pollen transcriptome, compared with the anther transcriptomes, possesses a higher proportion of sense transcripts (with TPM ≥5) with corresponding NATs (Table II).
Table II.
Numbers of antisense transcripts with and without their sense transcripts
The antisense transcripts are categorized according to their abundance in TPMs. For antisense transcripts with corresponding sense transcripts, their percentages of all sense transcripts are shown.
| Transcripts | Developing Anthers
|
Pollen | |||||
|---|---|---|---|---|---|---|---|
| Meiosis | Tetrad | Early Vacuolated | Late Vacuolated | Mitosis | Mature Pollen | ||
| No. of antisense transcripts (% of antisense transcripts) | |||||||
| TPM ≥5 | 1,510 (12.0) | 1,142 (16.5) | 1,761 (19.0) | 1,096 (16.4) | 756 (16.5) | 695 (17.8) | 790 (24.9) |
| TPM ≥10 | 785 (8.0) | 537 (11.3) | 804 (12.5) | 529 (11.2) | 429 (13.3) | 386 (15.0) | 495 (23.6) |
| TPM ≥100 | 56 (4.0) | 41 (5.7) | 63 (7.2) | 59 (7.4) | 57 (10.7) | 48 (10.3) | 103 (18.9) |
| TPM ≥1,000 | 2 (4.2) | 3 (6.3) | 3 (7.7) | 4 (9.1) | 6 (10.9) | 8 (11.9) | 13 (11.7) |
| No. of antisense transcripts without sense transcripts | |||||||
| TPM ≥5 | 394 | 487 | 480 | 481 | 351 | 283 | 184 |
| TPM ≥10 | 250 | 225 | 238 | 221 | 202 | 155 | 109 |
| TPM ≥100 | 16 | 18 | 21 | 29 | 26 | 15 | 10 |
| TPM ≥1,000 | 0 | 0 | 0 | 0 | 3 | 2 | 1 |
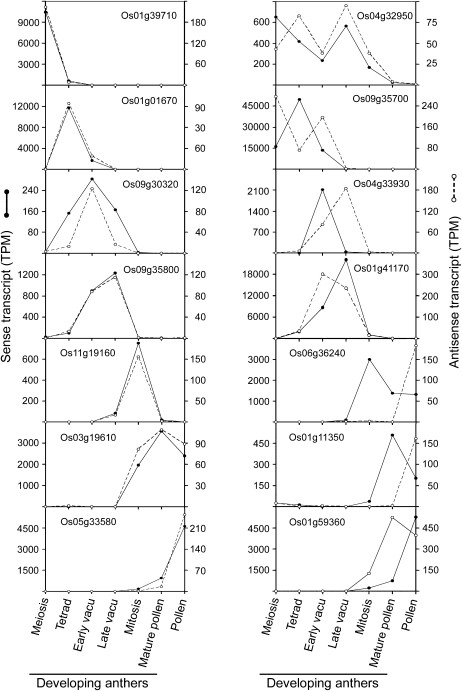
During anther development, the pattern of increase, peak, and decrease in level of some NATs matched that of their corresponding sense transcripts (Fig. 3, left column). However, many other NATs peaked in level before or after that of their corresponding sense transcripts (Fig. 3, right column). The occurrence of both matching and nonmatching developmental profiles of individual sense-antisense pairs may reflect that the antisense regulations are exerted via different mechanisms, such as transcriptional interference, RNA masking, different dsRNA-dependent mechanisms, and effect on DNA methylation (Lapidot and Pilpel, 2006). Regardless, in all cases, the levels of NATs are only approximately 10% to 50% of their corresponding sense transcripts, with the high 50% level usually reserved for sense transcripts with low TPM (Fig. 3).
Figure 3.
Variations in the developmental profiles of selected sense and antisense transcripts of various genes in anthers and pollen. Sense and antisense transcripts of an individual gene might share (left column) or not share (right column) the same developmental profiles. vacu, Vacuolated.
NATs that do not have the corresponding sense transcripts are also present in the anther transcriptomes but are substantially less abundant than NATs with their corresponding sense transcripts (Table II). The functions of these NATs without corresponding sense transcripts remain to be elucidated.
Anther Transcripts Largely Represented Those in Sporophytic Cells during Early Development But in Gametophytic Pollen during Late Development
A rice anther includes four layers of sporophytic cells enclosing a locule in which microspores mature (Fig. 1). It also includes some but not abundant vascular cells. A transcript in an anther transcriptome could be present in the sporophytic cell layers or in the gametophytic microspores, or both. Few transcripts are known to be present exclusively in young microspores and not in mature pollen or the sporophytic cells; UDP-Glc pyrophosphorylase may be such a microspore-specific transcript (Chen et al., 2007). Most transcripts in mature pollen first appear in microspores and increase in amount until maturation (Suen et al., 2003). Thus, an in silico subtraction of pollen transcripts from anther transcripts would yield transcripts that are largely restricted to the sporophytic anther, especially the tapetum with dense cytoplasm (Fig. 1). We performed this subtraction by obtaining anther transcripts that are >5× in TPM than are pollen transcripts and retained about 30% to 50% of all anther transcripts (Table I). A reversed subtraction to retain pollen transcripts that are >5× in TPM than are anther transcripts (averaged in the six stages) yielded microspore/pollen-specific transcripts (Table I).
Further in silico subtractions were performed to yield transcripts that are highly specific for sporophytic cells not just in anthers but also in the whole plant. We obtained transcripts in anthers that are >5× in TPM than are transcripts in pollen and other rice organs (Table I). Also, a reversed subtraction to retain pollen transcripts that are >5× in TPM than are transcripts in anthers (averaged in the six stages) and other rice organs yielded transcripts that are highly specific for microspores/pollen (Table I).
We examined the most abundant anther-minus-other-organs transcripts, which should be highly specific for anther sporophytic cells in a whole plant. Sample results of these transcripts of the tetrad-stage anther are shown in Table III, which lists the top transcripts according to their TPM, as well as their TPM in the transcriptomes of anthers of other developmental stages, pollen, and other organs. Several of the top transcripts have extremely high TPM (49K to 11K; i.e. each representing 5% to 1% of total mRNA). Most of these top transcripts have TPM >10× to 100× than those in all other organs, even though our in silico subtraction retained anther transcripts that are only >5× in TPM than those in pollen and other organs. Nine of the top 12 transcripts are completely absent in all other organs (TPM = 0).
Table III.
The most abundant transcripts in tetrad-stage anthers with TPM >5× those in pollen (current SBS transcriptomes) and other organs (previously reported MPSS transcriptomes) and the available mutants
Immature panicles contained developing flowers, including maturing anthers; thus, their transcripts were not considered in the 5× selection. The most abundant 50 transcripts in each developmental stage are shown in Supplemental Table S1, which includes full names of the mutant sources.
| TIGR_Gene Annotation | SBS
|
MPSS
|
Insertion Mutant | Activation Mutant | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Developing Anthers
|
Pollen | Germinated Seed | Young Leaf | Young Root | Mature Leaf | Mature Root | Stem | Immature Panicle | Ovary and Stigma | Pollen | |||||||||
| Meiosis | Tetrad | Early Vacuolated | Late Vacuolated | Mitosis | Mature Pollen | ||||||||||||||
| TPM | |||||||||||||||||||
| Os09g35700 | Lipid-transfer p | 16,153 | 49,751 | 13,603 | 81 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 708 | 0 | 0 | ||
| Os03g59380 | Lipid-transfer p | 1,358 | 17,792 | 20,728 | 35,555 | 512 | 169 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 399 | 29 | 3 | TRIM | |
| Os01g03670 | Dihydroflavonol-4-red | 47 | 11,738 | 1,649 | 16 | 5 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 117 | 0 | 0 | ||
| Os01g41710 | Chlorophyll a-b bind p | 1,373 | 11,472 | 17,209 | 17,289 | 2,617 | 45 | 0 | 29 | 20 | 0 | 0 | 0 | 0 | 369 | 55 | 0 | PFG, TRIM | |
| Os01g12020 | Lipid-transfer p | 9,549 | 9,683 | 105 | 29 | 19 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 120 | 0 | 0 | TRIM | |
| Os02g45530 | GMC oxidoreductase | 131 | 7,496 | 3,672 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | TRIM | TRIM |
| Os08g43290 | Lipid-transfer p | 554 | 6,768 | 1,261 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3,537 | 0 | 0 | TRIM | |
| Os07g46210 | Unknown p | 2 | 5,127 | 2,210 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | TRIM | |
| Os10g42620 | Dihydroflavonol-4-red | 9 | 4,613 | 2,445 | 345 | 7 | 1 | 1 | 22 | 0 | 0 | 0 | 0 | 8 | 26 | 3 | 0 | PFG, RMD, TRIM | TRIM |
| Os04g08280 | Unknown p | 2 | 3,728 | 342 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 119 | 0 | 0 | ||
| Os04g27950 | Unknown p | 16 | 3,015 | 273 | 128 | 33 | 9 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 15 | 0 | 0 | PFG, RMD | TRIM |
| Os01g14850 | MFS18 p | 1,237 | 2,407 | 426 | 88 | 10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | TRIM | |
| Os02g44630 | Aquaporin | 477 | 2,391 | 1,496 | 883 | 421 | 685 | 0 | 149 | 112 | 29 | 0 | 51 | 346 | 245 | 51 | 70 | PFG, RTIM | |
| Os03g47910 | Unknown p | 807 | 2,357 | 923 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 489 | 0 | 0 | ||
| Os08g03520 | Gly-rich p | 1,782 | 2,108 | 1,598 | 3,136 | 206 | 66 | 2 | 47 | 15 | 0 | 0 | 72 | 87 | 69 | 167 | 0 | ||
| Os01g41170 | Unknown p | 0 | 1,914 | 8,675 | 22,039 | 1,105 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 318 | 0 | 242 | PFG | |
| Os01g49650 | Lipid-transfer p | 2,632 | 1,850 | 39 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1,591 | 0 | 0 | Geno, PFG | |
| Os08g03682 | Cytochrome P450 | 43 | 1,705 | 812 | 5 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 469 | 0 | 0 | ||
| Os02g38260 | Hydrolase | 0 | 1,628 | 96 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 0 | 0 | ||
Table III also reveals the developmental changes of the levels of the transcripts. For many of the transcripts, their levels peaked at the tetrad stage (as so selected) or the immediately preceding or following stage. Some transcripts peaked in level at relatively late development (e.g. Os01g59380 and Os01g41170 peaking at the late-vacuole stage). Lists of the top 50 transcripts whose TPM peak at each of the six progressive developmental stages can be found in Supplemental Table S1.
A High Proportion of the Anther-Minus-Pollen Transcripts Were Restricted to the Tapetum
The anther-minus-pollen transcripts in meiosis- to late-vacuolated-stage anthers (Table I) could be largely those restricted to the tapetum, which has outstandingly dense cytoplasm among all sporophytic cells. We did not test these transcripts for tapetum specificity with use of in situ RNA hybridization, which is semiquantitative at best, and because of the wide occurrence of antisense transcripts (described in a preceding section), the hybridization results would be even less reliable. Instead, we explored the literature for reported tapetum-located transcripts in various plant species and tested whether rice orthologs of their genes encode transcripts in our anther-minus-pollen transcriptomes. Among the approximately 50 transcripts reported to be present in the tapeta of various plant species, some are of closely related paralogs and some do not have related rice ortholog transcripts (e.g. Brassicaceae oleosin genes; Kim et al., 2002). We filtered these results into 23 rice genes with ortholog transcripts present specifically in tapeta of other species. We then examined the expression patterns of these rice genes in the anther-minus-pollen transcriptomes and transcriptomes of other rice organs (Supplemental Table S2). Strikingly, all of the 23 transcripts are present in our anther-minus-pollen transcriptomes. In addition, 13 of the 23 transcripts are absent in all other rice organs. The remaining 10 transcripts are also present at various levels in other rice organs. Overall, the analytical results strongly support our notion that a very high proportion of the anther-minus-pollen transcripts are restricted to the tapetum. Nevertheless, some of these transcripts may be present in young but not mature microspores or other non-tapetum sporophytic cells such as those of the outermost anther cell layer or lining the stomium. Future researchers would need to make additional explorations.
Earlier, we used laser dissection to obtain tapetum cells from anthers. The resulting tapetum sample did not yield good-quality RNA and, upon bulk production, might contain contaminants of other anther constituents. Also, we were unable to obtain dissected tapetum cells from anthers of defined progressive developmental stages. Thus, we resorted to using the subtraction procedure to obtain tapetum-specific transcripts.
DNA Insertion Mutants of Genes Encoding About 50% of the Largely Anther-Minus-Other-Organs Transcripts Are Available
The mutants include insertion mutants, with DNA (T-DNA or transposable element) inserted into the ORF or the projected 5′- or 3′-UTR of the gene, and activation mutants, with DNA (available as activator-T-DNA insertion mutants) inserted into the genes or an intergene region within a defined distance from adjacent genes. These mutants are available at the various rice mutant centers (Table III; Supplemental Table S1).
Approximately 25% of the Most-Abundant Anther-Minus-Other-Organs Transcripts Encode Secretory Proteins
We explored the use of the transcriptomes to define two major tapetum events of great biological significance, namely, the secretion at a global scale (this section) and the synthesis of exine and its extracellular transport (next section). In the secretion study, we used the program PSORT (http:///psort.nibb.ac.jp/form.html) to predict subcellular locations of proteins encoded by the anther transcripts. We focused on the anther-minus-other-organs transcripts (largely those of the tapetum) that encode extracellular proteins (i.e. secretory proteins). The results are shown in Table IV, which also includes data from the transcriptomes of pollen and other organs for comparison. The number of secretory proteins encoded by all the anther-minus-other-organs transcripts (TPM ≥10) was fairly similar (approximately 8% of all transcript-generated proteins) during the first three developmental stages but rapidly decreased thereafter. Within each of the early stages, the percentages of transcript-generated proteins that are secretory increased from approximately 8% to approximately 12% to approximately 30% with increasing transcript TPM from ≥10 to ≥100 to ≥1,000, respectively. The findings reveal that active secretion of the tapetum occurs largely during early development and that many of the secretory proteins are abundant. Such active secretion is less obvious in leaves and ovaries plus stigma but similar in pollen and mature roots (Table IV) that have to interact extensively with the cellular exterior.
Table IV.
Secretory proteins encoded by anther-sporophytic-specific transcripts of progressive developmental stages
Transcripts were selected for their TPM >5× those in other organs (see legends to Tables I and III). They were further chosen for encoding proteins present outside the cell (probability ≥0.5 PSORT score). These transcripts are categorized according to their TPM of ≥10, 100 and 1,000, and their percentages in the total transcripts are shown. Transcripts encoding outside proteins in MPSS transcriptomes of leaves, roots, and ovaries are included for comparison. The lower portion summarizes groupings of the most abundant secretory proteins according to their possible functions. A list of all the outside proteins from transcripts of TPM ≥100 of each developmental stage is provided in Table S3.
| SBS
|
MPSS
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Developing Anthers
|
Pollen | Mature Leaf | Mature Root | Ovary and Stigma | ||||||
| Meiosis | Tetrad | Early Vacuolated | Late Vacuolated | Mitosis | Mature Pollen | |||||
| No. of transcripts (% transcripts encoding exterior proteins) | ||||||||||
| Transcripts | ||||||||||
| TPM ≥10 | 1,403 (7.1) | 933 (7.6) | 1,147 (8.3) | 885 (5.8) | 426 (5.2) | 200 (4.5) | 437 (10.5) | 195 (6.7) | 594 (12.5) | 367 (5.2) |
| TPM ≥100 | 247 (12.6) | 248 (12.5) | 288 (11.5) | 262 (7.3) | 89 (9.0) | 31 (9.7) | 247 (13.8) | 78 (2.6) | 152 (10.5) | 141 (6.4) |
| TPM ≥1,000 | 22 (31.8) | 33 (27.3) | 28 (39.3) | 26 (15.4) | 13 (23.1) | 4 (0.0) | 83 (15.7) | 21 (4.8) | 15 (20.0) | 15 (20.0) |
| Exterior proteins encoded by transcripts with: | TPM ≥100 | TPM ≥1,000 | ||||||||
| Total in all stages | 73 | 25 | ||||||||
| Lipid transfer/related proteins | 9 (12.3%) | 8 (32.0%) | ||||||||
| Unknown/hypothetical proteins | 25 (34.3%) | 7 (28.0%) | ||||||||
| Hydrolases (including wall-related) | 20 (27.4%) | 7 (28.0%) | ||||||||
| Others | 19 (26.0%) | 3 (12.0%) | ||||||||
Many of the secretory proteins encoded by the anther-minus-other-organs transcripts of the 6 developmental stages are similar. After consolidation, we found 73 unique proteins encoded by transcripts with TPM ≥100 and 25 encoded by transcripts with TPM ≥1,000 (Table IV; Supplemental Table S3). These proteins were grouped according to their possible functions: lipid-transfer and lipid-related proteins, unknown/hypothetical proteins, hydrolases related to wall and other polymers, and a mixture of others. The lipid-transfer and lipid-related proteins and the unknown/hypothetical proteins encoded by transcripts of TPM ≥100 and ≥1,000 are the most abundant (Table IV) and essentially absent in the transcriptomes of pollen and other organs (Supplemental Table S3). Thus, massive secretion of these proteins is unique to the sporophytic anthers, mostly the tapetum.
The secretory proteins annotated as unknown/hypothetical proteins of undefined functions represent approximately 30% of the abundant secretory proteins (Table IV). This proportion is markedly higher than the 1.1% proteins annotated as unknown/hypothetical secretory proteins (probability ≥0.5 PSORT score) encoded by all rice genes. The findings reiterate that we are still far from understanding but can use the current transcriptome data to explore, the active secretory role the tapetum plays in microspore maturation.
Transcripts Encoding Proteins Potentially Related to Pollen Exine Formation Were Selected from the Anther Transcriptomes
We also used the transcriptomes to select transcripts encoding proteins potentially related to pollen exine formation. Exine components are synthesized in the tapetum, and the exine structure gradually appears on the microspore surface during early anther development. Exine is a complex polymer postulated to consist of OH-FAs and isopropanoids linked by ester and ether bonds. In plant cells, FAs are hydroxylated in the endoplasmic reticulum (ER) via Cyt P450 (Cabello-Hurtado et al., 1998; Kandel et al., 2006; Morant et al., 2007), and enzymes for isopropanoid synthesis also locate in the ER (Winkel, 2004). The formation of ester bonds in exine are likely catalyzed by acyltransferases, which may locate in the ER, as most acyltransferases do (Voelker and Kinney, 2001; Kim et al., 2005; D'Auria, 2006). Ether bonds in exine may be formed spontaneously (Holmgren et al., 2006).
We hypothesize that in the tapetum cell, OH-FAs and isopropanoids are synthesized and then linked with acyl and ether bonds to become exine intermediates in the ER. These intermediates are transferred from the ER to the locule via secretory vesicles. Along the subcellular secretory pathway and in the locule, the water-insoluble intermediates are stabilized by lipid transfer proteins (LTPs). This hypothesized mechanism is more complicated than that for cuticular wax and suberin synthesis (Pollard et al., 2008; Samuels et al., 2008), with additional involvements of isopropanoids and intercellular transport. We further hypothesize that aquatic plants evolved to land plants with the production of exine to protect the air-borne pollen and that the exine-synthesizing enzymes were initially, and many still are, restricted to the anthers, especially the tapetum. We looked for the following proteins encoded by anther-minus-pollen transcripts that peaked in level at or around the tetrad stage, when exine begins to appear on the microspores (Fig. 1): (1) P450 for OH-FA synthesis; (2) enzymes for isopropanoid synthesis; (3) acyltransferases; and (4) LTPs. All were found, except that transcripts encoding enzymes for isopropanoid synthesis are ubiquitous. Enzymes for isopropanoid synthesis, unlike those for the synthesis of other exine materials, likely evolved in diverse organs and tissues to produce various isopropanoid-related molecules for different physiological functions (Graham, 1998; Winkel-Shirley, 2001).
Table V shows the selected transcripts encoding P450s, acyltransferases, and LTPs. Most of the transcripts peaked in level at the tetrad stage, and these developmental changes were confirmed by RT-PCR (Fig. 2, B and C). The selected transcripts were essentially absent in the transcriptomes of pollen (Table V) and other rice organs (data not shown). The transcripts encoding P450s and acyltransferases do not have exceedingly high TPM, as is befitting proteins for catalysis. On the contrary, the transcripts encoding LTPs, proposed to play a carrier role, are highly abundant, in thousands or tens of thousands of TPM (together representing 5%–10% of the total mRNA). The program PSORT predicted that the P450s and acyltransferases locate in the ER or its subsequent secretory pathway (vesicles and plasma membranes) and that the LTPs are outside the cell, with one in the vacuole. All the above results are consistent with our hypothesized mechanism of exine biosynthesis.
Table V.
Transcripts encoding three groups of proteins putatively related to exine synthesis in sporophytic anthers of progressive developmental stages
Their TPM, the predicted subcellular locations of their encoded proteins (with probability ≥0.5 PSORT score), and the available DNA insertion mutants (see legend to Table III) are shown. Transcript of the gene Os11g37280, which does not have the restriction site for SBS analyses but encodes an LTP detected in the anther locule (to be described in Fig. 4), is included. RT-PCR analysis showed that the Os11g37280 transcript was present during development (indicated with ++++) in the tetrad and early-vacuolated stages (Fig. 2C). The peak TPM of each transcript in the various developmental stages is shown in bold.
| TIGR_Gene | Developing Anthers
|
Pollen | Predicted Subcellular Location | Insertion Mutant | Activation Mutant | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Meiosis | Tetrad | Early Vacuolated | Late Vacuolated | Mitosis | Mature Pollen | |||||
| TPM | ||||||||||
| Cyto P450 | ||||||||||
| Os01g63930 | 25 | 6 | 1 | 1 | 0 | 0 | 0 | PM | PFG | TRIM |
| Os04g48210 | 0 | 674 | 448 | 10 | 29 | 8 | 0 | ER | ||
| Os05g29750 | 3 | 9 | 0 | 0 | 0 | 0 | 0 | ER | PFG, RMD | |
| Os08g03682 | 43 | 1,705 | 812 | 5 | 1 | 0 | 2 | ER | ||
| Os09g28390 | 156 | 256 | 76 | 30 | 170 | 9 | 1 | ER | PFG, RMD | TRIM |
| UCD | ||||||||||
| Acyltransferases | ||||||||||
| Os01g26000 | 16 | 447 | 331 | 109 | 28 | 0 | 0 | PM | PFG, RMD | TRIM |
| Os02g56860 | 68 | 26 | 1 | 0 | 0 | 5 | 0 | ER | PFG | |
| Os08g44840 | 8 | 614 | 651 | 165 | 10 | 4 | 0 | Unknown | PFG, TRIM | TRIM |
| LTP | ||||||||||
| Os01g12020 | 9,549 | 9,683 | 105 | 29 | 19 | 8 | 0 | Outside | TRIM | |
| Os01g49650 | 2,632 | 1,850 | 39 | 9 | 0 | 0 | 0 | Outside | GP, PFG | |
| Os08g43290 | 554 | 6,768 | 1,261 | 28 | 0 | 0 | 0 | Outside | TRIM | |
| Os09g35700 | 16,153 | 49,751 | 13,603 | 81 | 1 | 4 | 1 | Outside | ||
| Os11g37280 | ++++ | ++++ | Vacuole | UCD | TRIM | |||||
The search was a success, because we were able to pinpoint several genes out of the numerous genes encoding P450 (>100 genes), acyltransferases (>100 genes), and LTPs (38 genes). Most of the candidate genes have available DNA insertion and activation mutants (Table V), and Arabidopsis mutants of ortholog genes also exist. The phenotypes of these mutants could be loss of exine and male sterility. The selected members within each gene family encoding P450s, acyltransferases, and LTPs are not closely similar (phylogenetic trees not shown). These low similarities reduce the probability of the selected proteins within each group carrying out redundant functions, and thus a knockout mutant of one of these proteins is likely to have the phenotype of exine-less pollen and even male sterility. Whether exine-less pollen leads to male sterility in inbreeding species such as rice and Arabidopsis, in which fertilization occurs within the same closed flower, is unknown. One of the selected P450s (Os08g03682) has an Arabidopsis ortholog (At01g01280) recently shown to encode a P450 that hydroxylates FA for exine formation; the Arabidopsis gene transcript is restricted to flowers, and the knockout mutant has impaired pollen-wall development and partial male sterility (Morant et al., 2007).
LTPs and their related proteins have eight conserved Cys residues and a defined overall structure (Arondel et al., 2000; Jose-Estanyol et al., 2004). They are encoded by members of a gene family; rice and Arabidopsis have 38 and 70 genes, respectively. All LTPs have an N-terminal ER-targeting signal peptide for the secretory pathway. They carry out different functions, one of which could be for lipid transfer to the cellular exterior. Several earlier studies showed that LTP transcripts were present in the anther tapetum and/or microspores (e.g. Lauga et al., 2000; Matsuhira et al., 2007). The current study defines the expression of specific LTP genes during anther development, the massive levels of the transcripts, and their putative functions.
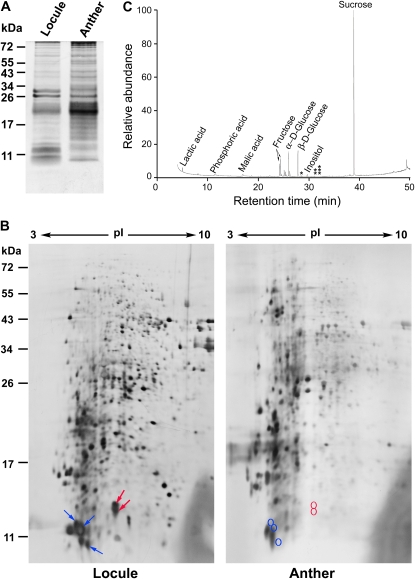
Some of the Abundant LTPs Were Detected in the Anther Locule Fluid, Which Had No Free FAs or OH-FAs
Locule fluid from anthers of the tetrad to early-vacuolated stages obtained via micropipetting was analyzed (Fig. 4). We first checked the locule fluid sample for contaminants, which would have arisen from other parts of the anthers during the micropipetting. We compared the protein patterns of the sample with that of a sample of total anther extract via SDS-PAGE (Fig. 4A). The two samples shared some proteins of apparently similar molecular masses, but the locule fluid sample was highly enriched with proteins of 10 to 13 kD. The high enrichment suggests that the locule fluid sample was not severely contaminated with proteins from other anther cells. The locule sample and the total anther sample were further separated into numerous spots of proteins via two-dimensional (2-D) gel electrophoresis. We paid attention to the abundant 10- to 13-kD proteins, which could be LTPs of low molecular masses. In the locule fluid sample (Fig. 4B), three closely situated spots were identified via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALTI-TOF) as being those of an LTP (from Os01g12020); the several spots could represent different isoforms, one isoform in combination with different types of SDS micelles, one isoform modified differently during treatment with urea, polypeptides resulted from mild proteolysis occurred naturally or artificially during sample handling, or different phosphorylated forms. Two other closely located spots were identified as being those of another LTP (from Os11g37280). All these protein spots derived from Os01g12020 and Os11g37280 were highly enriched in the locule fluid sample as compared with those in total anther extract (Fig. 4B). The two LTPs were identified in a preceding section (Table V) to be potential carriers of exine precursors. Several other protein spots of low-molecular mass proteins on the 2-D gel could not be positively identified. They could represent proteins of the abundant LTP encoded by Os09g35700 (Table V), which does not have appropriate trypsin sites for easy MALTI-TOF identification, and the less-abundant LTPs encoded by Os01g49650 and Os08g43290 (Table V). If these LTPs were not present in the locule, it could be because they: (1) were selectively degraded into amino acid for absorption into the microspores; (2) were actively recycled back to the tapetum cells; (3) remained in the secretory pathway within the tapetum cells and never traveled to the locule; or (4) became components of the exine structure on the microspore surface.
Figure 4.
Proteins and small metabolites in an anther locule fluid sample. The locule fluid sample was obtained from tetrad/early-vacuolated-stage anthers via micropipetting. A, One-dimensional PAGE gel of the locule sample and anther total extract. Proteins of 10 to 13 kD were highly enriched in the locule sample. Positions of molecular mass are shown on the left. B, 2-D PAGE gels of the two samples. In the locule fluid sample, three blue arrows indicate the proteins identified as LTP encoded by Os01g12020, and two red arrows depict LTP encoded by Os11g37280; the corresponding positions of the two LTPs in the total anther extract are circled. C, GC/MS separation and identification of small metabolites in the locule sample. One, two, and three stars indicate the elution times of markers palmitic, oleic, and stearic acids, respectively.
Small metabolites in the locule fluid sample were analyzed with gas chromatography/mass spectrometry (GC/MS; Fig. 4C), which could detect molecules smaller than 1 kD but not larger molecules such as the hypothetical exine intermediates. The most abundant metabolites resolved were Suc, Glc, and Fru, which presumably were sugars transported from the tapetum to the locule and then the microspores. No free FA or OH-FA was detected. The absence of free FAs and OH-FAs is consistent with, although does not validate, our hypothesis that FAs are hydroxylated and esterified in the ER in the tapetum cells rather than in the locule as exine monomers and intermediates, which are then transported to the locule and the microspore surface.
CONCLUSION
The current anther transcriptomes are substantially more advanced than those reported previously. The major advances include the comprehensiveness and quantification of transcripts, the finite division of the anthers into progressive developmental stages, the available data on antisense transcripts, and the compatibility of the anther transcriptomes with existing rice MPSS transcriptomes. Because of these advances, we made considerable progress in mining data related to two major tapetum events of great biological significance. First, we were able to select all the secretory proteins uniquely produced by the tapetum, which allows for interpretation of the tapetum secretory function and its shift during development. Knowledge of these proteins being restricted to the tapetum and having specific developmental profiles, plus the availability of mutants, allows for immediate functional exploration. An unusually high proportion of the abundant tapetum secretory proteins, as compared with secretory proteins derived from the whole rice genome, are annotated as having undefined functions; these functions now can be explored. Second, we were successful in selecting from hundreds of transcripts several transcripts encoding Cyt P450, acyltransferases, and LTPs as potential proteins for lipid exine synthesis and transport during anther development. The findings provide immediate opportunities to dissect the century-old mystery of the structure and synthesis of the extremely chemical-resistant exine.
Much more information than what has been presented in this report and the online version is available at the Web site http://rice.sinica.edu.tw/rice anther for researchers to dissect other functions of the tapetum and other anther constituents. For example, information in the transcriptomes can be used to generate male sterility for the production of hybrid seed. Most commercial hybrid seed production has relied on reversible deleterious genes expressed in the diploid tapetum. The active promoters (producing transcripts of high TPM) of many of the genes whose transcripts are completely restricted to the tapetum at an early developmental stage may be used, along with an added reversible signal motif, to drive a deleterious ORF to generate diploid male sterility for hybrid seed production.
MATERIALS AND METHODS
Plant Material
Plants were grown in the field from rice (Oryza sativa subsp. japonica ‘Tainung 67’) seed, kindly provided by Taiwan Agriculture Research Institute. Developing anthers were obtained from panicles. The youngest panicles were those with the auricle of the flag leaf 2 cm above the auricle of the preceding leaf. The oldest panicles were those with a 3- to 5-cm protrusion from the flag leaf sheath (details shown in Supplemental Table S4). Anthers were divided into six developmental stages according to the size/color of spikelets and anthers (Fig. 1), which were named according to cytological changes in the microspores used by some other laboratories (Li et al., 2006). Pollen was collected from panicles with about one-half of the spikelets dehisced. Samples were fixed for microscopy or frozen in liquid nitrogen and stored at −80°C for RNA extraction.
RNA Extraction and SBS Transcriptome Construction and RT-PCR
Anthers and pollen were ground with MagNa Lyser Green Beads (Roche), and their RNA was extracted with TRIzol reagents (GibcoBRL) for construction of SBS transcriptomes and RT-PCR. In the construction of SBS transcriptomes by Solexa, mRNA was converted to dscDNA with use of oligo(dT) primers (and thus all the eventually obtained sense and antisense sequences were derived from poly-A-containing cDNA). The cDNAs were digested with Sau3AI and ligated to MmeI site-containing adapters. They were cleaved with MmeI to release 21- to 22-bp fragments each containing a Sau3AI site. The fragments were ligated to sequencing adopters at both ends and randomly fixed into the inside surface of flow cell channels. For amplification and bridging, all four labeled reversible terminators (A, C, G, and T), primers, and DNA polymerase were added into the flow cells, and the resulting fragments were paired to matched DNA fragments. After laser excitation, the image of emitted fluorescence from each cluster on the flow cell was captured to identify the nucleotide base for each cluster. This synthetic cycle was repeated 16 times to obtain a 20-mer signature (a signature began with GATC and so the synthetic cycle was repeated only 16 times). Signature annotation was preformed through complete 20-mer matching to RAP2 (http://rapdb.dna.affrc.go.jp/) and TIGR ver.5 annotation databases (http://rice.plantbiology.msu.edu/). For RT-PCR, first-strand cDNA was synthesized with an oligo(dT)20 primer and then subjected to PCR with primers (Supplemental Table S5) as described (Suen et al., 2003). PCR products were separated on 1.0% agarose gels containing ethidium bromide and visualized and photographed on top of a UV light.
Bioinformatics Analysis
Two criteria were used to select 20-mer sequenced signatures for further analyses: reliable signatures of at least 5 TPM in each transcriptome, and a unique signature (i.e. hitting the rice genome sequence only once to avoid confusion among paralogs that have identical short sequences; Nobuta et al., 2007). A sense transcript is the sequence corresponding to an ORF and 500 bp in the 3′-UTR, and its complementary sequence is that of an antisense transcript. Subcellular locations of proteins encoded by transcripts were predicted with the software PSORT package (http://psort.ims.u-tokyo.ac.jp/).
Protein Analysis
Locule fluid from anthers of tetrad to early vacuolated stages was micropipetted (15 μm diameter) and immediately mixed 1:1 (v:v) with a solution of 50 mm NaPO4, 10 mm EDTA, and 1% dithiothreitol for one-dimensional SDS-PAGE (16% acrylamide). For 2-D gel electrophoresis, the locule fluid was mixed 1:1 (v/v) with a solution of 8 m urea, 2% CHAPS, and 1% dithiothreitol. The locule fluid collected from about 600 anthers and containing about 5 μg protein was mixed with ampholytes (pH 3–10) and loaded onto an immobilized pH gradient strip (7 cm, nonlinear, pH 3–10, Bio-Rad). After electrophoresis, the strip was subjected to SDS-PAGE (12.5% acrylamide). The protein spots were visualized with Forum Silver staining, and the proteins were subjected to trypsin digestion and then MS (MALTI-TOF) analysis with Voyager DE-STR (PerSeptive Biosystems) according to the manufacturer's instructions.
Metabolite Analysis
Metabolites in locule fluid from approximately 360 anthers, obtained as described, were converted to trimethylsilyl derivatives with N,O-bis-trimethylsilyl-trifluoroacetamide in 1% trimethylchlorosilane and heating at 70°C for 60 min. GC/MS analyses were performed in a ThermoFinniganTrace GC 2000 attached to a Polaris Q mass detector with a Xcalibur software system. The sample was injected into a 30-m × 0.25-mm (i.d.) × 0.25-μm (film thickness) Rtx-5MS fused silica capillary column chemically bonded with a 5% diphenyl-95% dimethylpolysiloxane cross-linked stationary phase (Restek). The injector and ion source temperatures were 230°C and 200°C, respectively. The oven temperature program was 80°C for 5 min, increasing at 5°C/min to 300°C, and then 300°C for 1 min. Helium was used as the carrier gas at 1 mL/min. The mass spectrometer was operated in electron impact mode (70 eV). Data acquisition was performed in the full scan mode from mass/charge of 50 to 650 with a scan time of 0.58 s. GC/MS-detected peaks were identified by comparing both the MS spectra and the retention index with compounds in libraries (NIST by Wiley, http://www.wiley.com/WileyCDA/WileyTitle/productCd-0470425172.html) and reference compounds available commercially.
Sequence data from this article can be found in http://rice.sinica.edu.tw.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Table S1. Top 50 transcripts in anther-minus-other-organs transcriptomes of various developmental stages.
Supplemental Table S2. Tapetum-specific transcripts in anther-minus-pollen transcriptomes.
Supplemental Table S3. Abundant transcripts encoding secretory proteins from anther-minus-other-organs transcriptomes.
Supplemental Table S4. Characteristics of anther samples
Supplemental Table S5. Primers used in RT-PCR.
Supplementary Material
Acknowledgments
We sincerely thank Drs. Wann-Neng Jan and Tuan-Nan Wen, Institute of Plant and Microbial Biology, Academia Sinica, for electron microscopy and proteomics, respectively, Dr. Chia-Chin Hou, Agricultural Biotechnology Center, Academia Sinica, for GC/MS, and Dr. Ming-Shen Lai, Taiwan Agricultural Research Institute, for collection of rice pollen.
This work was supported by Academic Sinica (a pilot grant and a Vice President Special Fund) and by the U.S. Department of Agriculture-National Research Initiative (grant no. 2005–02429).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anthony Huang (anthony.huang@ucr.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Ahlers F, Lambert J, Wiermann R (2003) Acetylation and silylation of piperidine solubilized sporopollenin from pollen of Typha angustifolia L. Z Naturforsch [C] 58 807–811 [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol 145 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai M, Ariizumi T, Endo M, Hatakeyama K, Kuwata C, Shibata D, Toriyama K, Watanabe M (2003) Identification of anther-specific genes in a cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex Plant Reprod 15 213–220 [Google Scholar]
- Arondel V, Vergnolle C, Cantrel C, Kader JC (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci 157 1–12 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Hempel FD, McCormick S, Zambryski P (2000) Reproductive development. In Buchanan BB, Gruissem W, Jones RL, eds, Biochemistry and Molecular Biology of Plants. American Society for Plant Biologists, Rockville, MD, pp 988–1043
- Bishop R, Shah T, Pelle R, Hoyle D, Pearson T, Haines L, Brass A, Hulme H, Graham SP, Taracha EL, et al (2005) Analysis of the transcriptome of the protozoan Theileria parva using MPSS reveals that the majority of genes are transcriptionally active in the schizont stage. Nucleic Acids Res 33 5503–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174 483–498 [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Batard Y, Salaun JP, Durst F, Pinot F, Werck-Reichhart D (1998) Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J Biol Chem 273 7260–7267 [DOI] [PubMed] [Google Scholar]
- Carlsson J, Lagercrantz U, Sundstrom J, Teixeira R, Wellmer F, Meyerowitz EM, Glimelius K (2007) Microarray analysis reveals altered expression of a large number of nuclear genes in developing cytoplasmic male sterile Brassica napus flowers. Plant J 49 452–462 [DOI] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2001) Plant biology. Pollen tube guidance: right on target. Science 293 1441–1442 [DOI] [PubMed] [Google Scholar]
- D'Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9 331–340 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TL (1998) Flavonoid and flavonoid glycoside metabolism in Arabidopsis. Plant Physiol Biochem 36 135–144 [Google Scholar]
- Hesse M, Pacini E, Willemse M (1993) The Tapetum: Cytology, Function, Biochemistry and Evolution. Springer-Verlag, Wein, New York, pp 1–152
- Holmgren A, Brunow G, Henriksson G, Zhang L, Ralph J (2006) Non-enzymatic reduction of quinine methides during oxidative coupling of monolignols: implications for the origin of benzyl structures in lignins. Org Biomol Chem 4 3456–3461 [DOI] [PubMed] [Google Scholar]
- Jose-Estanyol M, Gomis-Ruth FX, Puigdomenech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42 355–365 [DOI] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Olry A, Diss L, Benveniste I, Pinot F (2006) Cytochrome P450-dependent fatty acid hydroxylases in plants. Phytochem Rev 5 359–372 [Google Scholar]
- Kang J, Zhang G, Bonnema G, Fang Z, Wang X (2008) Global analysis of gene expression in flower buds of Ms-cd1 Brassica oleracea conferring male sterility by using an Arabidopsis microarray. Plant Mol Biol 66 177–192 [DOI] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AH (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277 22677–22684 [DOI] [PubMed] [Google Scholar]
- Kim HU, Li Y, Huang AH (2005) Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y (2006) Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep 7 1216–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga B, Charbonnel-Campaa L, Combes D (2000) Characterization of MZm3-3, a Zea mays tapetum-specific transcript. Plant Sci 157 65–75 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Duncan D, Morrow DJ, Fernandes J, Walbot V (2007) Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell types. Plant J 50 637–648 [DOI] [PubMed] [Google Scholar]
- Ma J, Morrow DJ, Fernandes J, Walbot V (2006) Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Biol 7 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik MR, Wang F, Dirpaul JM, Zhou N, Polowick PL, Ferrie AM, Krochko JE (2007) Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol 144 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko H, Endo M, Saito H, Hakozaki H, Park JI, Kawagishi-Kobayashi M, Takada Y, Okabe T, Kamada M, Takahashi H, et al (2006) Anther-specific genes, which expressed through microsporogenesis, are temporally and spatially regulated in model legume, Lotus japonicus. Genes Genet Syst 81 57–62 [DOI] [PubMed] [Google Scholar]
- Matsuhira H, Shinada H, Yui-Kurino R, Hamato N, Umeda M, Mikami T, Kubo T (2007) An anther-specific lipid transfer protein gene in sugar beet: its expression is strongly reduced in male-sterile plants with Owen cytoplasm. Physiol Plant 129 407–414 [Google Scholar]
- Morant M, Jorgensen K, Schaller H, Pinot F, Moller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al (2007) An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25 473–477 [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Kaushik S, Sakai H, Arteaga-Vazquez M, Sanchez-Leon N, Ghazal H, Vielle-Calzada JP, Meyers BC (2008) A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol 8 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13 236–246 [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59 683–707 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16 S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Wu SS, Chang HC, Dhugga KS, Huang AHC (2003) Cell wall reactive proteins in the coat and wall of maize pollen: potential role in pollen tube growth on the stigma and through the style. J Biol Chem 278 43672–43681 [DOI] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52 335–361 [DOI] [PubMed] [Google Scholar]
- Wang Z, Liang Y, Li C, Xu Y, Lan L, Zhao D, Chen C, Xu Z, Xue Y, Chong K (2005) Microarray analysis of gene expression involved in anther development in rice (Oryza sativa L.). Plant Mol Biol 58 721–737 [DOI] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J 52 14–29 [DOI] [PubMed] [Google Scholar]
- Winkel BS (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55 85–107 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.