Genomic resources have significantly impacted plant biology research in recent years. Cell biology has been further enabled by an ongoing revolution in visualization technologies. Using fluorescent proteins (FPs), we now have unprecedented views of cellular architecture, and we can study real-time dynamics of cell structure, function, and protein localization. To date, these technologies have been most widely used in Arabidopsis (Arabidopsis thaliana); however, the grasses provide a unique opportunity to study the underlying mechanisms and inter-related controls of cell growth, morphogenesis, and physiology in leading crop models.
Here, we present a resource that leverages the emerging maize (Zea mays) genome sequence to develop tools to study protein structure and function in a cellular context. Traditionally, such studies relied on fixed tissue or FP fusions driven by constitutive promoters, which can lead to significant artifacts. The maize genome sequence now provides access to regulatory regions that can be used to drive native expression. We have developed streamlined methods to generate maize FP-tagged lines using these regulatory elements, allowing analysis of tissue-specific expression and localized function. Identification of diverse proteins that function in specific subcellular compartments will provide the tools for understanding basic developmental, biochemical, and physiological processes in maize, with direct application potential for crop improvement.
METHODOLOGY
We developed a protocol to generate fusion proteins with yellow (YFP), cyan (CFP), or red (RFP) color variants of FPs driven by native regulatory elements, based on our previous work in Arabidopsis (Tian et al., 2004). In brief, the method uses triple template PCR to generate products of the full genomic sequence with the FP insert, which is flanked by linker peptides to minimize folding interference between the FP and tagged protein. The product is cloned using the Gateway system (Invitrogen) into the donor vector, pDONR207. The tagged gene is transferred into binary destination vectors and ultimately transformed into maize. Full details of the protocols are available at http://maize.jcvi.org/cellgenomics/protocol/maizeTTprotocolGFP_111405.shtml.
Candidate genes were selected for tagging based on several criteria, including, as first priority, the availability of full genomic sequence plus regulatory regions that included 3 kb upstream and 1 kb downstream of the coding region. A size limit of 8 to 9 kb for the full genomic region with the FP insertion is imposed to ensure good cloning efficiency. Given these size constraints, we next prioritized genes that encoded proteins with robust predicted functions. These decisions were based on homology to other well-studied proteins, known localizations to specific compartments, and/or corroborating antibody or expression data. Genes with available mutations were also given high priority so as to provide a means of functional complementation. Our final criterion was to include candidates that would label the full range of cellular compartments.
Approximately 25% of genes tagged were derived from specific requests that met our designated tagging criteria from researchers in the maize community. The remaining 75% of genes were selected initially by searching approximately 2,500 TIGR (The Institute for Genomic Research) Release 5 AZMs (Assembled Zea mays; Chan et al., 2006) that were >5 kb and 300 TIGR maize bacterial artificial chromosome sequences that were available at the start of the research project. Fgenesh gene predictions (Salamov and Solovyev, 2000) and homology to ESTs and protein databases were generated and subcellular localization predicted using PSORT (protein sorting tool; Nakai and Kanehisa, 1992). Candidate gene sequences were also selected from GenBank, Maize Assembled Genomic Island sequences (Fu et al., 2005), ChromDB (Gendler et al., 2007), or MaizeGDB (Lawrence et al., 2007). Agrobacterium-mediated transformation of maize was performed at the Iowa State University Plant Transformation Facility (Frame et al., 2002; Paz et al., 2006). Plantlets were screened for FP expression and seeds bulked by crossing plants to the inbred line B73.
DESCRIPTION OF SELECTED TAGGED LINES
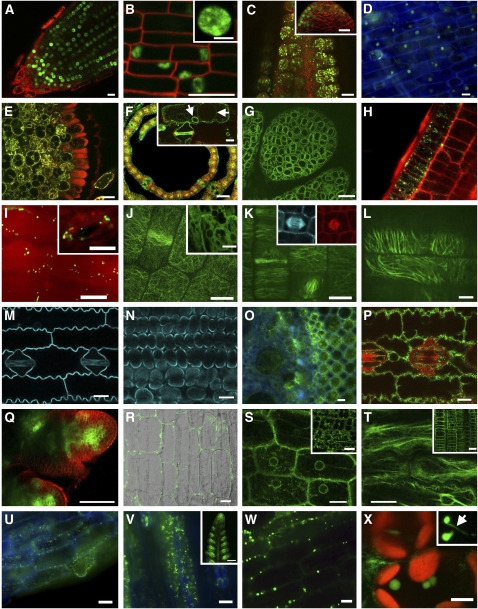
Figure 1 depicts a subset of images representative of the tagged lines generated. The tagged lines mark most compartments in maize cells for use in diverse research programs. For example, proteins localized to the nucleus are ideal as constitutive markers and for studying chromosome dynamics and developmental regulation (Fig. 1, A–D). Here we display two nuclear markers, each serving a unique function. Histone H1 is a linker histone that functions in maintaining higher order chromatin structure and in epigenetic regulation (Misteli et al., 2000). Ten H1 genes have been identified in maize (www.Chromdb.org), and we selected HON110 for tagging. The YFP-tagged protein localized to nuclei in roots, leaves, and inflorescences (Fig. 1, A–C, respectively). We observed punctate subnuclear foci representative of heterochromatin (Misteli et al., 2000). Interestingly, labeling was low in the central region of meristems, likely reflecting the reduced activity and/or cell cycle of these cells (Fig. 1C, inset).
Figure 1.
Sample images of markers for cell compartments. Nuclear markers include Histone H1, HON110 in root tip (A), leaf (B), and inflorescence apex (C) and MRE11B in expanding husk (D). Vacuole membrane marker: TIP1 in root (E), leaf (F), and shoot apex (G). Peroxisome marker: PEX11 in root (H) and leaf (I). Tubulin cytoskeleton markers: α-TUB1 in leaf (J–L) and root (J, inset) and β-TUB1 in spindles (K, insets). Plasma membrane marker: PIP2-1 in leaf epidermis (M) and mesophyll (N). Cell wall marker: EXP1 in root (O), leaf (P), and inflorescence apex (Q). Vesicle trafficking markers: ROP7 in root (R) and RAB2A1 in leaf (S). Actin cytoskeleton marker: FIM in leaf (T) and root (T, inset). Mitochondrial marker: HSP22 in root (U), leaf (V), and inflorescence apex (V, inset). Plastid marker: LOX10 in leaf (W and X). (See text for gene abbreviations and Web site for additional images; http://maize.jcvi.org/cellgenomics/index.shtml.) The images are confocal (A–C, E–L, R–T, W, and X) or wide-field micrographs. Red color represents tissues counter-stained with FM4-64 (A–C, E, H, P, and Q), RFP expression (K, inset), or chlorophyll autofluorescence (F and X). Blue color represents cell wall autofluorescence (D, O, U, and V) or CFP expression (K [inset], M, and N). Scale bars = 20 μm, except B inset, X = 5 μm; K = 8 μm; C, F, Q, and V inset = 100 μm.
The second nuclear marker displayed is Meiotic Recombination11B (MRE11B) tagged with YFP. MRE11 proteins are tools for studying chromosome repair, due to their interactions with RAD50 and NBS1, which mediate double-stranded break repair and recombination (Waterworth et al., 2007). MRE11B-YFP was observed in maize nuclei, consistent with transient heterologous expression assays of the NBS1 partner in dicots (Waterworth et al., 2007). MRE11B-YFP localized strongly in dividing and growing tissues of the shoot and root tip (data not shown) and expanding leaf and husk cells (Fig. 1D), consistent with its hypothesized DNA repair function. This tagged protein will be useful for comparative study of meiotic recombination in the grasses (Lohmiller et al., 2008).
As a marker for the vacuole, we tagged a maize aquaporin, tonoplast intrinsic protein1 (TIP1; Barrieu et al., 1998). TIP1-YFP localized to vacuolar membranes in root, leaf, and inflorescence spikelet primordia (Fig. 1, E–G, respectively; also see F, inset, vacuole-like structures in an expanding leaf cell, arrowed), as well as reticulate and perinuclear structures typical of endoplasmic reticulum. In general, TIP1-YFP was broadly expressed. Interestingly, the fusion protein was less abundant in root epidermal cells, with the exception of the root hairs (Fig. 1E). This marker will be useful for studies of vacuole organization and function and in the response of plants to various stresses.
Peroxin11 (PEX11) was tagged as a marker for peroxisomes, small organelles that perform a variety of oxidative cellular functions. PEX11 is targeted to peroxisomes in Arabidopsis and has been implicated in peroxisome proliferation (Lingard et al., 2008). The PEX11-YFP fusion localized to small, highly motile organelles that resemble peroxisomes in root and leaf cells (Fig. 1, H and I), including guard cells (Fig. 1I, inset). This marker will be useful for studies of cellular oxidative processes and organelle dynamics (a movie can be seen at http://maize.jcvi.org/cellgenomics/index.shtml).
FP tags of tubulin (TUB) are essential for studying in vivo cytoskeletal dynamics and organization. Because α-TUB1 and β-TUB1 dimerize during polymerization, we tagged α-TUB1 with YFP (Fig. 1, J–L) and β-TUB1 with CFP or RFP (Fig. 1K, insets) to provide multiple FP tools for studying microtubule (MT) dynamics and array formation. All expected MT arrays associated with cell division were observed in leaf, root, and inflorescence apices, including preprophase bands (Fig. 1J), spindles (Fig. 1K), and phragmoplasts. Additionally, cortical arrays in YFP-tagged α-TUB1 leaf cells show typical random orientation (Fig. 1J) during cell division and more aligned orientation during expansion, observations that are uniquely possible in the oriented cells of the grasses. Interestingly, β-TUB1-tagged MTs were observed only in dividing cells (Fig. 1K, insets), consistent with prior studies showing β-Tub1 gene expression is downregulated in nondividing tissues (Hussey et al., 1990). Although YFP-tagged α-TUB1 was highly expressed in roots, its incorporation into MTs appeared lower than in shoots (Fig. 1J, inset).
Immediately adjacent to the cortical array of MTs lies the plasma membrane (PM) compartment that serves as a gate between the cytoplasm and the cell wall. We marked this compartment by fusing CFP to a PM intrinsic protein (PIP2-1), an integral membrane aquaporin that transports water, small uncharged solutes, and gases across the PM (Chaumont et al., 2001). PIP2-1-CFP localized to the PM in epidermis (Fig. 1M) and mesophyll (Fig. 1N), as expected based on known PIP2 function in other systems. PIPs are generally regulated by drought stress in dicots (Mahdieh et al., 2008), suggesting our PIP2-1-CFP line could be tested as a physiological marker of water transport and drought responses in maize. We recently published another marker for PM, YFP-tagged PINFORMED1 that functions as an auxin efflux carrier and will be a useful reporter of maize auxin responses (Gallavotti et al., 2008).
α-Expansin1 (EXP1) contributes to cell growth during wall extension and stress relaxation (Sampedro and Cosgrove, 2005). Of the five α-EXP genes identified in maize (Wu et al., 2001), EXP1 was tagged due to its reported expression along the growth gradient of a developing maize leaf (Muller et al., 2007). EXP1-YFP appeared correctly targeted to cell walls (Fig. 1P), as confirmed in plasmolysis experiments (data not shown). The fusion protein was also abundant in the cytoplasm of root and leaf (Fig. 1, O and P), particularly prior to cell morphogenesis (data not shown). Inflorescence apices counterstained with FM4-64 (red) show EXP1-YFP localized to sites of incipient primordia (Fig. 1Q). Our observations support the idea that EXP1 function (i.e. for cell wall loosening) is one of the earliest structural events in emergence of an organ primordium (Pien et al., 2001).
Vesicle trafficking is a highly coordinated system of intracompartmental transport in eukaryotes, mediated by small GTPases in the Ras superfamily. ROPs and RABs are two subfamilies that have diverged extensively in plants (Christensen et al., 2003; Zhang et al., 2007) and may have acquired new plant-specific functions despite highly conserved protein structure across eukaryotes (Zhang et al., 2007). Maize ROP7 was tagged with YFP and localized only to root cells in punctate structures as well as being dispersed in the cytoplasm (Fig. 1R). RAB2A1, tagged with YFP, localized across cellular compartments in young dividing and expanding leaf and root cells (Fig. 1S). It is interesting to note that RAB2A1-YFP was restricted to subsidiary cells in mature leaf (data not shown), similar to the down-regulation of expression at the leaf tip observed in maize and sorghum (Sorghum bicolor; Zhang et al., 2006).
The actin cytoskeleton is a dynamic structure associated with cell division, expansion, organelle movement, and maintenance of subcellular compartments. Study of actin in plant cells has been largely limited to fixed tissue, possibly due to the failure of actin filament assembly when subunits are tagged. However, the actin-binding protein fimbrin permits in vivo observation of actin cytoskeleton through its direct binding to intact microfilaments (Wang et al., 2008). Through bioinformatic identification of a maize homolog to Arabidopsis fimbrin, we fused a double YFP to the actin-binding domain 2 of maize fimbrin and present here initial observations of T0 seedlings, showing localization to a filamentous network in leaf and root cells (Fig. 1T and inset).
Mitochondrial markers are important tools to study organelle morphology, development, and function. Heat shock proteins (HSPs) are useful because they can be either conditional or constitutive (Shemetov et al., 2008), thus providing experimental control of expression. We tagged HSP22, a molecular chaperone, with YFP and the protein was constitutively localized in mitochondria in maize leaf, root, and inflorescence apices (Fig. 1, U and V). Expression of HSP18, also tagged with YFP, was induced by thermal stress (data not shown). Thus, these tagged proteins will be useful as conditional or constitutive mitochondrial markers for developmental study.
Lipoxygenase10 (LOX10) is thought to function in synthesis of oxylipins, including jasmonic acid, which functions in defense responses as well as in plant development (Nemchenko et al., 2006). LOX10-YFP localized to organelles that lacked standard chlorophyll autofluorescence (Fig. 1, W and X) and were smaller than mature chloroplasts. These organelles may be leucoplasts or other intermediary plastids, an idea supported by the observation of stromule-like structures (Fig. 1X, inset) that are characteristic of plastids (Kohler et al., 1997). This marker will be useful to study plastid biogenesis and to understand compartmentalization of the oxylipin pathways.
MATERIAL AVAILABILITY, UTILITY IN RESEARCH, AND INSIGHTS INTO FUTURE DEVELOPMENT
Currently, approximately 40 proteins have been tagged with FP and T2 generation seeds are available by request through our Web site at http://maize.jcvi.org/tigr-scripts/maize/cellgenomics/seed_request.pl. We aim to continue tagging up to 100 proteins for compartments not yet labeled and for proteins developmentally regulated, physiologically controlled, and conditionally expressed. We will leverage advances in live cell imaging techniques and continue to mine the emerging maize genome sequence for new regulatory regions and genes of interest. In addition to their value for cell and developmental studies, these FP-tagged proteins will be useful for proteomic analysis of protein complexes and for protein-protein interaction studies. Our constructs are designed for easy replacement of the FP tag with any other tag, allowing flexibility in future experiments. We are also developing additional cloning techniques to increase throughput. The techniques described here could easily be applied to other grasses, providing a broad-based cell biological resource to the plant biology community and serving to provide new views into protein and cellular imaging.
Acknowledgments
We thank Dr. R. Tsien for his generous gift of citrine YFP and mRFP1 plasmids. We thank Tim Mulligan for excellent plant care at Cold Spring Harbor Laboratory, Pete Gallins and Ryan Pendleton for greenhouse assistance at the University of Wyoming, and Kan Wang, Bronwyn Frame, and the staff of the Iowa State University Plant Transformation Facility for outstanding maize transformation services. A.W.S. thanks Dr. Mike Tamkun (Colorado State University) for use of live cell imaging facilities and Dr. Laurie Smith (University of California, San Diego) for use of a spinning disc confocal microscope.
This work was supported by the National Science Foundation (DBI no. 0501862).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anne W. Sylvester (annesyl@uwyo.edu).
References
- Barrieu F, Thomas D, Marty-Mazars D, Charbonnier M, Marty F (1998) Tonoplast intrinsic proteins from cauliflower (Brassica oleracea L. var. botrytis): immunological analysis, cDNA cloning and evidence for expression in meristematic tissues. Planta 204 335–344 [DOI] [PubMed] [Google Scholar]
- Chan AP, Pertea G, Cheung F, Lee D, Zheng L, Whitelaw C, Pontaroli AC, SanMiguel P, Yuan Y, Bennetzen J, et al (2006) The TIGR Maize Database. Nucleic Acids Res 34 D771–D776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125 1206–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen TM, Vejlupkova Z, Sharma YK, Arthur KM, Spatafora JW, Albright CA, Meeley RB, Duvick JP, Quatrano RS, Fowler JE (2003) Conserved subgroups and developmental regulation in the monocot rop gene family. Plant Physiol 133 1791–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, et al (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102 12282–12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008) The relationship between auxin transport and maize branching. Plant Physiol 147 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler K, Paulsen T, Napoli C (2007) ChromDB: the chromatin database. Nucleic Acids Res 36 D298–D302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Haas N, Hunsperger J, Larkin J, Snustad DP, Silflow CD (1990) The beta-tubulin gene family in Zea mays: two differentially expressed beta-tubulin genes. Plant Mol Biol 15 957–972 [DOI] [PubMed] [Google Scholar]
- Kohler RH, Cao J, Zipfel WR, Webb WW, Hanson MR (1997) Exchange of protein molecules through connections between higher plant plastids. Science 276 2039–2042 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC (2007) MaizeGDB's new data types, resources and activities. Nucleic Acids Res 35 D895–D900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN (2008) Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmiller LD, De Muyt A, Howard B, Offenberg HH, Heyting C, Grelon M, Anderson LK (2008) Cytological analysis of MRE11 protein during early meiotic prophase I in Arabidopsis and tomato. Chromosoma 117 277–288 [DOI] [PubMed] [Google Scholar]
- Mahdieh M, Mostajeran A, Horie T, Katsuhara M (2008) Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant Cell Physiol 49 801–813 [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408 877–881 [DOI] [PubMed] [Google Scholar]
- Muller B, Bourdais G, Reidy B, Bencivenni C, Massonneau A, Condamine P, Rolland G, Conejero G, Rogowsky P, Tardieu F (2007) Association of specific expansins with growth in maize leaves is maintained under environmental, genetic, and developmental sources of variation. Plant Physiol 143 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 0888–7543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemchenko A, Kunze S, Feussner I, Kolomiets M (2006) Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J Exp Bot 57 3767–3779 [DOI] [PubMed] [Google Scholar]
- Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25 206–213 [DOI] [PubMed] [Google Scholar]
- Pien D, Wyrzykowska J, Fleming A (2001) Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J 25 663–674 [DOI] [PubMed] [Google Scholar]
- Salamov A, Solovyev V (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemetov A, Seit-Nebi A, Gusev N (2008) Structure, properties, and functions of the human small heat-shock protein HSP22 (HspB8, H11, E2IG1): a critical review. J Neurosci Res 86 264–269 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB (2008) Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol 177 525–536 [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Altun C, Armstrong SJ, Roberts N, Dean PJ, Young K, Weil CF, Bray CM, West CE (2007) NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J 52 41–52 [DOI] [PubMed] [Google Scholar]
- Wu Y, Meeley RB, Cosgrove DJ (2001) Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol 126 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hill DR, Sylvester AW (2007) Diversification of the RAB guanosine triphosphatase family in dicots and monocots. J Integr Plant Biol 49 1129–1141 [Google Scholar]
- Zhang J, Sylvester AW, Li DQ, Sun XP (2006) Complementation and expression analysis of SoRab1A and SoRab2A in sugarcane demonstrates their functional diversification. J Integr Plant Biol 48 1450–1457 [Google Scholar]