Abstract
The apoplast of plant cells, which carries out multiple functions in plant metabolism and signaling, is not only a barrier but also the linker between the environment and the protoplast. To investigate the role of apoplastic proteins in the salt stress response, 10-d-old rice (Oryza sativa) plants were treated with 200 mm NaCl for 1, 3, or 6 h, and the soluble apoplast proteins were extracted for differential analysis compared with untreated controls using two-dimensional electrophoresis. Ten protein spots that increased or decreased significantly in abundance were identified by mass spectrometry. These proteins included some well-known biotic and abiotic stress-related proteins. Among them, an apoplastic protein, with extracellular domain-like cysteine-rich motifs (DUF26), O. sativa root meander curling (OsRMC), has shown drastically increased abundance in response to salt stress during the initial phase. OsRMC RNA interference transgenic rice has been generated to assess the function of OsRMC in the salt stress response. The results show that knocking down the expression level of OsRMC in transgenic rice led to insensitive seed germination, enhanced growth inhibition, and improved salt stress tolerance to NaCl than in untransgenic plants. These results indicate that plant apoplastic proteins may have important roles in the plant salt stress response.
Salt stress is one of the most significant abiotic stresses and affects every aspect of plant physiology and metabolism. During salt stress, Na+ enters the cells and accumulates to a concentration that induces ionic and osmotic stress in plants. Plant cells respond and adapt to these adverse conditions through signaling networks (Lee et al., 2004). Understanding the signaling pathway of plant salt resistance is important for improving plant salt tolerance, especially for improving agricultural productivity in irrigated land.
To study the signaling network of plant salt adaptation, the most important thing is identification of the components involved. Much effort has been made to discover components or elements of signaling pathways involved in plant salt stress responses. Of all the molecular components and signaling pathways known so far, the best understood signaling pathway in salt ionic stress is the SOS pathway (Zhu, 2002). In addition to the SOS pathway, other signaling pathways and components have also been suggested to be involved in salt osmotic stress signaling. Several plant protein kinases have been found to be activated by osmotic stress (Liu et al., 2000; Zhu, 2002). Osmotic stress can increase the transcription levels of a number of protein kinase genes, including genes for a two-component His kinase, mitogen-activated protein kinase kinase kinase, mitogen-activated protein kinase kinase, and mitogen-activated protein kinase (Munnik et al., 1999; Mikolajczyk et al., 2000; Mizoguchi et al., 2000). Also, membrane phospholipids make up a dynamic system that generates a multitude of signaling molecules in addition to serving important structural roles during stress responses (Munnik et al., 2000; DeWald et al., 2001; Zhu, 2002). Recently, a new Ca2+-dependent membrane-binding protein (AnnAt1) involved in salt stress was identified by a proteomic approach (Lee et al., 2004). To date, most identified salt stress resistance-related signaling components have been localized to the cytosol or cell membrane. Although it is an important component of the plant cell, the plant apoplast has been ignored in studies of the salt stress response.
The apoplast is the portion of the plant cell outside the cell membrane. This region includes the cell walls and intercellular space of the plant (Dietz, 1997). The apoplast, which plays important roles in regulating plant physiological and developmental processes, is not only a barrier but also a linker between the environment and the protoplast; there are many organic and inorganic molecules, as well as enzymes, present in the plant apoplast. These molecules have crucial functions in plant cell metabolism (Nielsen and Schjoerring, 1998), including responses to pathogen stress (del Carmen Córdoba-Pedregosa et al., 2003; Misas-Villamil and van der Hoorn, 2008), cell division and proliferation, cell differentiation (Takeda et al., 2003), and, especially, responses to drought and salt stress (Brune et al., 1994; Dietz, 1997; Ramanjulu et al., 1999). Recently, there has been increasing evidence that plants use apoplastic peptide signals to regulate different plant developmental and other physiological processes, such as the systemin (Pearce et al., 1991), phytosulfokines (Matsubayashi and Sakagami, 1996), CLAVATA3 (Clark et al., 1997), and S-locus Cys-rich protein (Schopfer et al., 1999). A bioinformatics approach was taken to find putative secreted peptides in Arabidopsis (Arabidopsis thaliana); 33,809 open reading frames (ORFs) were deduced to code for putative secreted peptides, and according to microarray information, many of them may be expressed (Lease and Walker, 2006). To date, however, biological functions have been reported for only a few apoplastic proteins. These proteins, compared with the number of putative secreted proteins predicted by bioinformatics, are just the tip of the iceberg. Furthermore, the sequencing of the Arabidopsis and rice (Oryza sativa) genomes has revealed large numbers of putative receptor-like kinase genes. The potential ligands of many of these putative receptors have been predicted to be peptides. It is very likely that some of these receptor-like protein kinases may be involved in plant perception of, and response to, biotic and/or abiotic stress signals (Haffani et al., 2004). These data suggest that there may be a number of proteins in the plant apoplast that are involved in the plant stress response. The identification of these proteins during the initial phase of salt stress will be an important step toward understanding the role of these apoplastic proteins in the plant salt stress response.
The sequencing of many plant and animal genomes has revealed the fact that the regulation of a cell's biological activities mostly occurs at the level of protein degradation, interactions, and posttranslational modification. Proteomics is now emerging as a powerful tool for studying these protein dynamics, especially in plant stress responses (Salekdeh et al., 2002; Lee et al., 2004). A number of studies have been performed using proteomic approaches to identify pathogen response proteins in rice and Arabidopsis (Kim et al., 2003; Ndimba et al., 2003) and salt stress response proteins in the rice microsome (Lee et al., 2004), root (Yan et al., 2005), and leaf sheath (Abbasi and Komatsu, 2004). Dani et al. (2005) used a proteomic approach to analyze the proteome changes in the tobacco (Nicotiana tabacum) leaf apoplast during long-term (20 d) salt stress. Nevertheless, few studies have addressed the changes in the apoplast proteome in response to salt stress, especially during the initial phase of salt stress. In this study, the changes in the rice root apoplast proteins during the initial phase of salt stress were carefully investigated by two-dimensional electrophoresis (2-DE). Eight salt stress-regulated apoplast proteins were identified, including a secreted protein with extracellular domain-like Cys-rich motifs (DUF26) that has been reported to be involved in O. sativa root meander curling (OsRMC; Jiang et al., 2007). In our work, the roles of OsRMC in the response to salt stress have been revealed. OsRMC was up-regulated at the transcriptional and translational levels during the initial phase of salt stress. Our findings showed that knocking down the expression level of OsRMC resulted in more resistance to salt stress in transgenic rice.
RESULTS
Apoplastic Extracts Are Free of Cytosolic Contamination
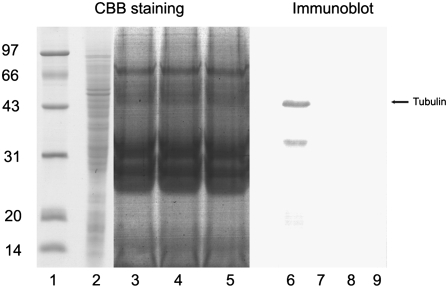
The vacuum infiltration method was used to obtain apoplastic fluid from the rice root. Because the procedures of vacuum infiltration and centrifugation could damage cells and contaminate apoplastic extracts with cytosolic ingredients, we used malate dehydrogenase (MDH) activity assay and immunobloting against tubulin to evaluate the quality of apoplastic extracts. Our results show that MDH activity could not be detected in our apoplast extract (Table I). To eliminate the possibility that the difference in MDH activity between the total cell extracts sample and the apoplastic extracts sample was due to the protein concentration difference, the total cell extract was diluted to a concentration that was similar to that of the apoplastic extracts and analyzed again for MDH activity (Table I); the normalized MDH activity in total cell extracts was even higher than in the undiluted samples. In addition, apoplastic extracts from three independent extraction procedures and soluble cell total extracts were separated and probed with the anti-tubulin monoclonal antibody. With an apparent molecular mass of 49 kD, tubulin was detectable in whole cell extracts but not in any of our apoplastic extracts (Fig. 1). These results proved that the apoplast extract samples used in this study were free of cytosolic contamination.
Table I.
MDH enzyme assay
Results shown present data collected from one independent experiment. Of all the MDH activities assayed in three independent experiments, the results were similar. One unit (U) was calculated using the change of 0.01 A340 per minute. ND, Not detected.
| Extract | Protein Concentration | Enzyme Activities | Relative Enzyme Activities |
|---|---|---|---|
| mg mL−1 | U mL−1 | U mg−1 | |
| Rice root total cell extracts | 0.65 | 180 ± 28 | 277 ± 43 |
| Diluted rice root total cell extracts | 0.065 | 38 ± 7 | 585 ± 108 |
| Rice root apoplastic extracts | 0.06 | ND | ND |
Figure 1.
Immunoblot analysis of apoplastic proteins and total soluble proteins. Proteins extracted from the apoplast (lanes 3, 4, 5, 7, 8, and 9) and total soluble protein (lanes 2 and 6), 10 μg lane−1, were stained with Coomassie Brilliant Blue R-250 (CBB; lanes 1–5) or transferred to a polyvinylidene difluoride membrane (lanes 6–9), which was probed with anti-α-tubulin monoclonal antibody. Lane 1 contains a protein molecular mass standard.
The Apoplastic Proteins Changed in Response to Salt Stress
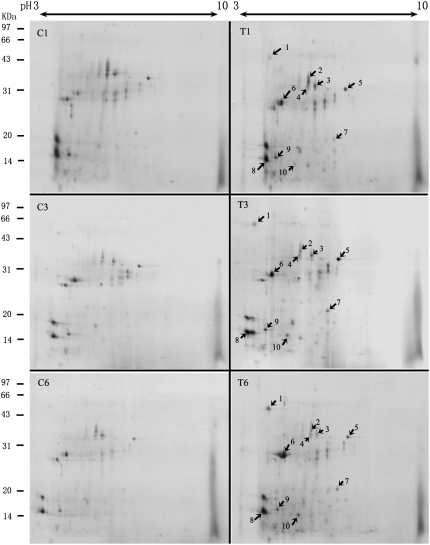
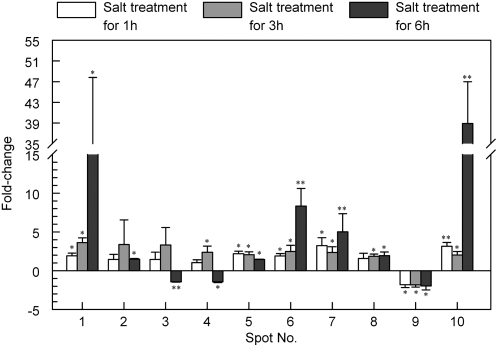
Rice root apoplast proteins from the 1-, 3-, and 6-h salt treatments and from the controls were analyzed by 2-DE. Approximately 100 spots from each sample were visualized by Coomassie Brilliant Blue staining (Fig. 2). Three independent experiments were conducted to ensure that the protein abundance changes at each time point were reproducible and significant. Software quantification showed that, although the protein expression profiles between the salt-stressed and untreated samples were similar to each other, the abundance of some spots changed significantly with salt stress treatment over time (Figs. 2 and 3). We found 10 spots that showed at least a 1.5-fold increase or decrease in abundance (P < 0.05) in response to salt stress (Figs. 2 and 3). Based on their expression profiles, these proteins were classified into three groups. The first group contains spots 1, 2, 5, 6, 7, 8, and 10, which showed a steady increase in abundance over the course of the treatment (Fig. 3). The second group contains spot 9 only, which showed a steady decrease in abundance across all treatment time points (Fig. 3). The third group contains spots 3 and 4. The abundance of the spots in this group increased at 3 h and decreased at 6 h of salt treatment (Fig. 3). The different expression patterns of the spots might imply different roles for these apoplastic proteins in plant salt stress responses.
Figure 2.
Protein expression profiles of the rice root apoplast after treatment for 1 h (C1 and T1), 3 h (C3 and T3), and 6 h (C6 and T6) with 200 mm NaCl (T1, T2, and T3) or regular medium (C1, C2, and C3). Arrows indicate the MALDI-TOF MS-identified protein spots whose abundance increased or decreased consistently following NaCl treatments.
Figure 3.
Histogram showing fold changes in abundance (following 200 mm NaCl treatment for 1, 3, and 6 h) for each of the protein spots in three independent experiments. Values plotted are mean values with sd. The significance of the differences between treated and untreated groups is indicated by asterisks: * P < 0.05, ** P < 0.01.
Identification of Salt Stress-Responsive Proteins by Mass Spectrometry
All 10 salt stress-responsive spots were subjected to in-gel digestion and analyzed by matrix-assisted laser-desorption ionization time of flight mass spectrometry (MALDI-TOF MS). The obtained peptide mass fingerprints were used to search the National Center for Biotechnology Information database using Mascot (http://www.matrixscience.com). The 10 identified proteins spots are listed in Supplemental Table S1. The experimentally calculated molecular mass and pI values of identified protein spots were consistent with the theoretical values, except for spots 1 and 9 (Supplemental Table S1).
To confirm that the proteins we identified were indeed apoplastic proteins, all identified protein sequences were searched using the TargetP program (www.cbs.dtu.dk/services/TargetP) for their predicted subcellular localization (Emanuelsson et al., 2000). As we can see from Supplemental Table S1, eight of 10 spots were predicted to be typical secretory proteins with signal peptide sequences.
OsRMC Is an Apoplastic Protein
Both spots 1 and 6 were identified as protein OsRMC (gi|19387274). To confirm the identities of spots 1 and 6, MS/MS was used for further analysis (Supplemental Figs. S1 and S2). The results showed that spots 1 and 6 represent the same protein, and the pI/molecular mass difference between the two spots may be due to posttranslational modification or dimerization. This protein has been further analyzed using bioinformatics tools. OsRMC belongs to the secreted type of DUF26 protein (Chen, 2001), with two extracellular domain-like Cys-rich motifs (DUF26). A sequence including 23 residues in the N terminus was predicted to be the signal peptide (http://www.cbs.dtu.dk/services/SignalP/; Nielsen et al., 1997; Moller et al., 2001; Supplemental Fig. S3).
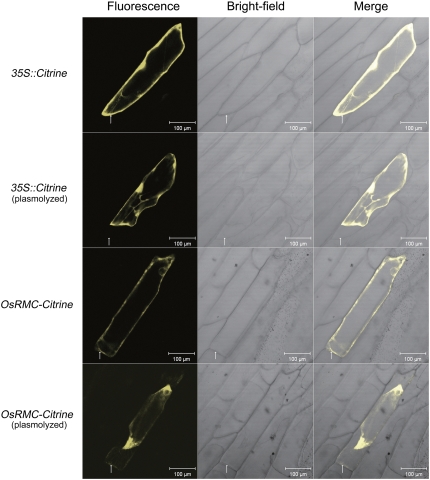
As OsRMC was obtained from an apoplastic sample and bioinformatics analysis also indicated that OsRMC is a putative secreted protein, we sought experimental evidence for its apoplastic localization. We used Citrine reporter gene, which encodes a new yellow fluorescent protein (YFP) that has lower pH sensitivity (Griesbeck et al., 2001), to determine the subcellular localization of OsRMC in vivo. The recombinant constructs of the 35S∷OsRMC-Citrine fusion gene were transiently expressed in onion (Allium cepa) epidermal cells by particle bombardment. Citrine expressed from the empty vector pAVA321 was used as a negative control. As shown in Figure 4, in the onion epidermal cell, OsRMC-Citrine was detected in the cell wall region after plasmolysis with 0.9 m mannitol, whereas for the empty vector control, the Citrine was not detected in the extracellular region but only throughout the intracellular region. This result confirms that OsRMC is an apoplastic protein.
Figure 4.
Subcellular localization of OsRMC transiently expressed in onion epidermal cells. The top two panels illustrate the empty vector control before and after the plasmolysis happened. The bottom two panels illustrate the construct containing OsRMC-Citrine before and after the plasmolysis happened. Citrine fluorescence, bright-field, and merged images are shown for each kind of transformation. Arrows indicate the cell wall region where plasmolysis happened. Bars = 100 μm. [See online article for color version of this figure.]
OsRMC Is a Salt Response Gene and Is Expressed in Different Developmental Stages
The 1,777-bp region upstream of the OsRMC start codon (ATG) was used to search the PLACE database (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999). Some putative abiotic stress response-related cis-acting elements were predicted in the OsRMC promoter sequence (Supplemental Fig. S3D). To determine the transcript level profile of the OsRMC gene in response to salt stress, the mRNA level of the OsRMC gene in response to salt stress was quantified using real-time quantitative PCR (Q-PCR) over a 12-h period. The results showed that, in 10-d-old rice seedlings, the mRNA of OsRMC was induced by the 200 mm NaCl treatment after 1 h, increased continuously to a peak at 6 h, and then started to decline by 12 h (Fig. 5B).
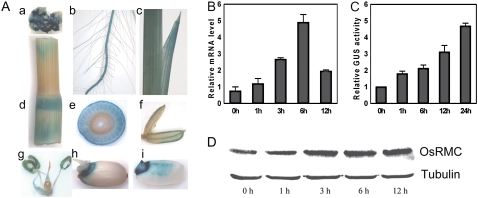
Figure 5.
Expression pattern of OsRMC in rice. A, Histochemical analysis of OsRMCpro∷GUS transgenic rice plants. Tissues from different stages were used for GUS staining: a, callus; b, seedling root; c, young leaf; d, stem; e, transverse section of a node; f, palea; g, stamen and pistil; h, germinated seed; i, vertical section of a germinated seed. B, Q-PCR analysis of OsRMC expression under salt stress treatment. Ten-day-old wild-type rice plants were treated with 200 mm NaCl for 1, 3, 6, and 12 h. Relative mRNA levels of OsRMC compared with those for untreated plants are shown. Actin was used as an internal control. Error bars indicate se of three replicates. C, Quantitative analysis of OsRMCpro∷GUS transgenic rice plants under salt stress. Ten-day-old transgenic rice plants were treated with 200 mm NaCl for 1, 6, 12, and 24 h. Relative GUS activities are shown compared with the GUS activity of untreated plants. Error bars indicate se of three replicates. D, Western-blot analysis of OsRMC expression at the protein level under salt stress treatment. The treatment condition was the same as for B. Tubulin was used as a loading control.
To investigate the transcriptional induction of the OsRMC gene in response to salt stress at the tissue level, transgenic rice lines that expressed the GUS reporter gene under the control of the OsRMC promoter were generated. The promoter was selected from the 1,777-bp sequence upstream of the ATG start codon. A reproducible expression pattern was found in three independent reporter lines. Under standard growth conditions, promoter activity was detected locally in the callus and was high in the roots and leaves of transgenic seedlings (Fig. 5A, a–c). In the mature rice plants, the stems and nodes, except the flag leaf, were stained blue (Fig. 5A, d and e). In the inflorescence, we could see GUS expression in the palea, lodicule, stamen, pistil, and especially in the anther (Fig. 5A, f and g). The seeds of transgenic rice were also stained (Fig. 5A, h and i). Expression of GUS was under the control of the OsRMC promoter in the transgenic plant, so the transcriptional induction of the OsRMC gene in response to salt treatment was determined by monitoring GUS activity. The GUS activity of OsRMCpro∷GUS plants in response to salt stress was measured over a 24-h period. Each data point is an average of three independent experiments. Treating the plants with 200 mm NaCl resulted in rapidly and continuously increased GUS activity (Fig. 5C).
To further confirm the protein level change of OsRMC in response to salt stress, we generated polyclonal antibodies against OsRMC. A western blot was used to confirm the induction of OsRMC at the translational level by salt stress. The results showed that the level of OsRMC protein increased when plants were treated with 200 mm NaCl for 1 h and kept increasing after 12 h under NaCl treatment (Fig. 5D). These results indicate that OsRMC is a salt stress-responsive gene and that salt stress promotes the expression of OsRMC at the transcriptional and the translational levels.
Knockdown of OsRMC Leads to Improved Salt Tolerance of Transgenic Plants
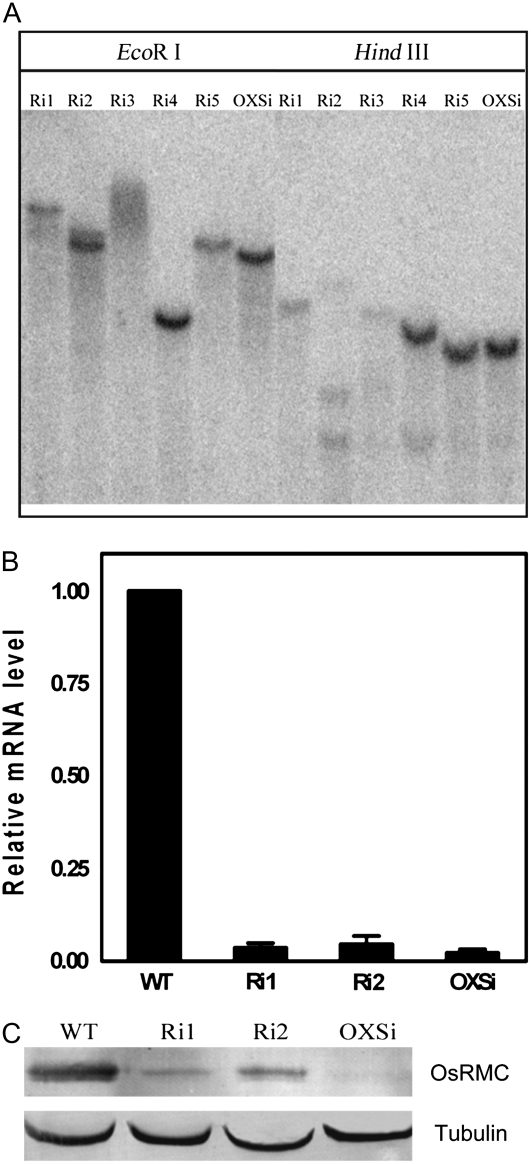
To assess the function of OsRMC in the salt stress response in vivo, an RNA interference (RNAi) construct (pTCK303) containing part of the coding sequence fragment (428 bp) to knock down OsRMC expression and an overexpression construct (pCAMBIA1300) containing the OsRMC full-length ORF (without the stop codon) fused to a GUS reporter gene driven by a ubiquitin promoter (Ubi-1pro∷OsRMC-Gus) were introduced into rice by Agrobacterium tumefaciens EHA105-mediated transformation. We examined the T-DNA insertions of select transgenic lines by Southern blot (Fig. 6A). Two independent RNAi homozygous lines (Ri1 and Ri2) and an overexpression homozygous line (OXSi) were used for further analysis. Real-time Q-PCR indicated the OsRMC mRNA expression levels in the RNAi lines were significantly knocked down. However, gene silencing of OsRMC also occurred in the overexpression line OXSi, resulting in a low OsRMC expression level, which was similar to that in the RNAi lines (Fig. 6B). To confirm the protein expression level for the RNAi lines, a western-blot assay was performed using an anti-OsRMC antibody. The results of the western blots matched those of the real-time Q-PCR: the protein expression level of OsRMC in the knockdown lines, including the overexpression silenced line, OXSi, was obviously reduced (Fig. 6C).
Figure 6.
Molecular identification of transgenic rice. A, Southern-blot analysis of independent RNAi and overexpression transgenic lines. Ri1 to Ri5 indicate RNAi lines; OXSi indicates an overexpression line. Hygromycin B was used as a probe for hybridization. B, Real-time Q-PCR analysis of OsRMC expression in knockdown lines. Relative mRNA levels of OsRMC compared with those for the wild-type rice (WT) are shown. Actin was used as an internal control. Error bars indicate se of three replicates. C, Western blot analysis of OsRMC expression at the protein level in knockdown lines. Tubulin was used as a loading control.
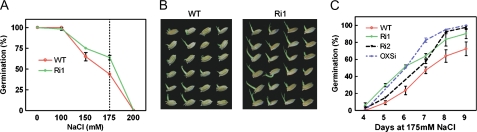
To analyze the functions of OsRMC under salinity stress, we tested the effect of NaCl on seed germinations of OsRMC knockdown lines. There were no differences in seed germination rates between the transgenic lines and the wild type under normal condition. We tried gradients of NaCl concentrations from 100 to 200 mm to treat seeds for 7 d and found that Ri1 seeds had a higher germination rate than the wild type, and in 175 mm NaCl treatment the difference was most distinct (Fig. 7, A and B). In 175 mm NaCl treatment, the seeds of Ri1, Ri2, and OXSi were soaked in NaCl solution on a plate containing a filter for 9 d. Seeds were scored for germination from the 4th d, based on whether or not the shoot length exceeded half of the seed length. In statistical analysis, the germination rates of the Ri1, Ri2, and OXSi transgenic lines were higher than that of the wild type in the presence of 175 mm NaCl (Fig. 7C).
Figure 7.
Insensitive seed germination to NaCl in OsRMC knockdown transgenic plants. A, Seed germination rates of knockdown line Ri1 and the wild type (WT) in gradient NaCl treatments for 7 d. Error bars indicate se of three replicates. B, Germinated seeds of Ri1 and the wild type in 175 mm NaCl after 9 d. C, Seed germination rates of knockdown lines in 175 mm NaCl. Error bars indicate se of three replicates. [See online article for color version of this figure.]
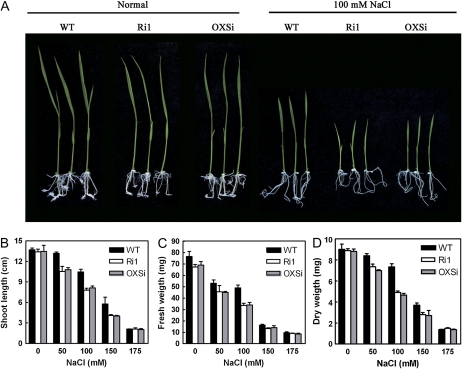
We also put the seeds of knockdown lines Ri1 and OXSi in half-strength Murashige and Skoog (1/2MS) medium containing 50 to 175 mm NaCl or without NaCl as a control to germinate and grow into seedlings. Ten-day-old seedlings were measured for shoot length, fresh weight, and dry weight of the shoot. The knockdown transgenic rice seedlings exhibited no difference in growth compared with wild-type rice seedlings under normal conditions, whereas in the presence of NaCl, the knockdown lines showed inhibited growth. In the 100 mm NaCl treatment, the difference was most distinct (Fig. 8A). Statistical analysis revealed that the knockdown plants exhibited distinct reduction in shoot length, fresh weight, and dry weight compared with control, wild-type plants (Fig. 8, B–D). In 175 mm NaCl, both the germinated seedlings of knockdown plants and control plants exhibited growth reduction with no apparent difference. The Ri2 knockdown line, which has a relatively higher expression level of OsRMC than Ri1 and OXSi (Fig. 6C), was also used for the growth assay in salt treatments and showed a similar phenotype of growth inhibition (data not shown).
Figure 8.
OsRMC knockdown lines show growth inhibition in NaCl treatments. A, Ten-day-old transgenic and wild-type (WT) rice plants grown in normal and 100 mm NaCl conditions. B to D, Shoot length (B), fresh weight (C), and dry weight (D) of seedlings in normal and gradient NaCl (50–175 mm) conditions. Error bars indicate se of three replicates. [See online article for color version of this figure.]
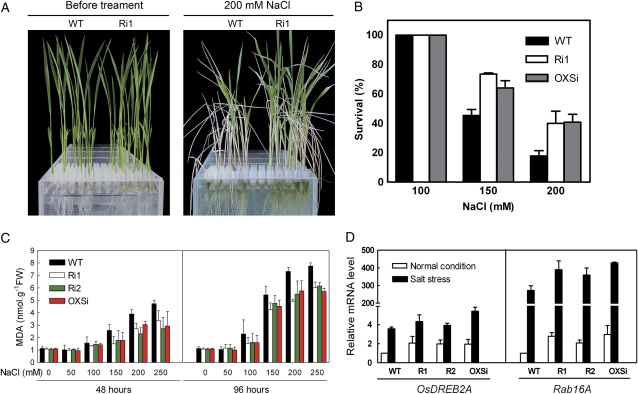
Growth rate reduction in the OsMRC-silenced plants could reflect either an adaptive response or an injury response of the plants under the stress (Zhu, 2001). In order to delineate these possibilities, we measured the seedling survival rates of OsRMC knockdown lines under salt stress conditions. For the survival assay, 10-d-old knockdown seedlings cultured in Hoagland liquid medium were transferred to Hoagland medium with gradient NaCl concentration from 100 to 200 mm for 4 d and then allowed to recover under normal conditions for 7 d, after which the survival rate was calculated. Plants that could not grow any more after the recovery period were considered to be dead. After recovering for 1 week, more transgenic seedlings survived than wild-type plants, which appeared mostly withered (Fig. 9A). The survival rates of Ri1 and OXSi were higher than that of the wild type after 7 d of recovery from 150 and 200 mm salt stress, whereas in recovery after 100 mm NaCl treatment both the knockdown lines and the wild type all survived (Fig. 9B). We measured the lipid peroxidation level in rice seedlings under salt stress, and the end product MDA contents of leaves were determined by thiobarbituric acid test (Heath and Parker, 1968). As shown in Figure 9C, MDA levels were elevated in both wild-type and transgenic plants within gradient NaCl treatments, but the degree of lipid peroxidation was significantly lower in knockdown plants than in wild-type plants, which means that the transgenic plants are more tolerant to stress injury.
Figure 9.
Knockdown of OsRMC leads to improved salt tolerance in transgenic plants. A, Growth of Ri1 before and after 200 mm NaCl treatment. B, Survival rates of knockdown lines in recovery after 100, 150, or 200 mm NaCl treatment. Error bars indicate se of three replicates. C, MDA contents of leaves from knockdown lines in gradient NaCl (50–250 mm) treatments for 48 and 96 h. Error bars indicate se of three replicates. FW, Fresh weight. D, Real-time Q-PCR analysis of the expression levels of salt-responsive genes OsDREB2A and Rab16A in wild-type and transgenic plants. Total RNA was extracted from leaves of 10-d-old plants grown under normal (white columns) or 200 mm NaCl (black columns) conditions for 6 h. Actin was used as an internal control. Error bars indicate se of three replicates. WT, Wild type.
The expression levels of two salt-responsive genes, OsDREB2A and Rab16A, also were monitored by real-time Q-PCR analysis. As shown in Figure 9D, both OsDREB2A and Rab16A showed higher expression in knockdown lines than in the wild type under normal conditions. When treated with 200 mm NaCl for 6 h, significant inductions of these two salt-responsive genes happened in all salt-treated plants, with higher expression levels in transgenic plants than in wild-type plants. Thus, knockdown of OsRMC results in up-regulated expression of OsDREB2A and Rab16A, which are involved in plant tolerance to salt stress.
DISCUSSION
To understand the signaling network underlying plant salt adaptation, a two-dimensional electrophoresis approach was used to identify the apoplastic components involved in this process. Despite the difficulties encountered in extracting and identifying proteins, apoplast proteomics has become an active field in recent years (Jamet et al., 2006, 2008). For a differential display analysis of apoplast proteins responding to salt stress, the quality of the apoplast-specific extract is crucial. There are several methods commonly used to extract the apoplast solution from the plant, including the application of pressure using a Scholander bomb, the vacuum perfusion method, the elution method, and the vacuum infiltration method (Lohaus et al., 2001; Jamet et al., 2008). Of all these methods, the vacuum infiltration method has been most widely used in obtaining apoplastic fluid from different plant species (Husted and Schjoerring, 1995; Lohaus et al., 2001; Dani et al., 2005). To avoid the cytoplasmic contamination of apoplast extracts, an MDH assay and a western-blot assay were used here to evaluate cytoplasmic contamination. MDH activity was detected in total protein extract controls but not in apoplastic extracts (Table I). The immunoblotting against tubulin indicated that tubulin was not present in apoplastic extracts but was present in total protein extracts (Fig. 1). Furthermore, all identified protein sequences were analyzed using the TargetP program (www.cbs.dtu.dk/services/TargetP) to predict their subcellular localization (Emanuelsson et al., 2000), since the sensitivity of mass spectrometry is 100 to 1,000 times greater than that of the biochemical assay (Jamet et al., 2006). As we can see from Supplemental Table S1, eight of 10 spots were predicted to be typical secretory proteins with a signal peptide sequence. These data indicate that our preparations of apoplastic proteins from the rice root did not show detectable intracellular contamination and therefore were appropriate for use in proteomic analysis.
Based on the 2-DE analysis, it was observed that the apoplastic proteins changed in response to salt stress during the initial phase. We found that protein expression pattern changes within 1 h (Figs. 2 and 3), which is quite a swift response. Previous studies had indicated that salt, drought, and heavy metals alter the apoplastic protein composition of leaves (Fuhrer, 1982; Brune et al., 1994; Dietz, 1997; Dani et al., 2005). According to the previous [35S]Met incorporation experiment (Dietz, 1997), apoplast protein was slightly changed after 1 h and continued to increase after 10 h of treatment with nickel. Previous work had also indicated that mRNA levels were altered in response to salt stress within 15 min in rice (Kawasaki et al., 2001). These results indicated that plants can rapidly respond and adapt to salt stress.
Ten significantly salt stress-responsive spots were subjected to in-gel digestion and analyzed by MALDI-TOF MS, and six kinds of protein were identified: OsRMC, a peroxidase, a glucanase, rab5B, a thioredoxin, and a putative pathogenesis-related protein. Based on their potential physiological functions, the salt stress-regulated apoplastic proteins identified in this study are all involved in the plant stress response. In plants, β-1,3-glucanase belongs to the same family of pathogenesis-related proteins as PR-2 and is strongly induced when plants respond to wounding or infection by fungal, bacterial, or viral pathogens (Trudel et al., 1998). Plant peroxidases are also well-studied PR proteins belonging to the PR-9 family (Swapan and Muthukrishnan, 1999). During the pathogenesis resistance response, apoplastic peroxides polymerize proteins and lignin or suberin precursors into plant cell walls (Young et al., 1995). This leads to the formation of a physical barrier that prevents the pathogen from penetrating the cell walls and spreading. Prb1 (a putative pathogenesis-related protein) contains a conserved domain named SCP, which is often seen in plant pathogenesis-related proteins (Marchler-Bauer et al., 2007) such as PR-1, a homolog of Prb1, and a salicylic acid (SA)-response marker gene widely used in SA and jasmonic acid (JA) response studies (Koornneef and Pieterse, 2008). Rab5B belongs to the Rab subfamily of the small G-protein superfamily. Rab GTPases are implicated in vesicle trafficking and are involved in multiple physiological processes in plants (Ueda et al., 2001; Preuss et al., 2004). In Mesembryanthemum crystallinum, a protein belonging to the plant rab5 family was regulated at the transcriptional level by early salt stress (Bolte et al., 2000). Also, thioredoxin reductase is part of the plant redox-regulating system, and there is evidence showing that thioredoxin participates in a number of physiological processes in plants, including abiotic and biotic stress responses, seed germination, self-incompatibility, cell division, and translation (Besse and Buchanan, 1997; Vieira Dos Santos and Rey, 2006).
It was interesting that spots 1 and 6 were both identified as an OsRMC, containing a predicted N-terminal signal peptide and two DUF26 domains (Supplemental Fig. S3C). Proteins with the DUF26 domain belong to a large protein family with many members in higher plants. All of these share similarities in the DUF26 domain (C-X8-C-X2-C, a Cys-rich repeat motif). These conserved Cys residues may function to maintain the three-dimensional structure and form a zinc finger motif to mediate protein-protein interactions or sense the redox changes in the extracellular space during plant defense responses (Chen, 2001). The majority of the DUF26 protein family consists of two types. One type is receptor-like kinases with the extracellular domain, the transmembrane domain, and the intracellular kinase domain, whereas the other type is secretory proteins, which have only the extracellular domain with Cys-rich motifs (DUF26). In the previous study of Jiang et al. (2007), the subcellular localization of an OsRMC-GFP fusion protein was analyzed in the epidermal cells of hypocotyls of transgenic Arabidopsis, and the results showed that the fusion protein was localized in the plasma membrane. As to our work, we have generated an OsRMC-Citrine fusion protein that was transiently expressed in onion epidermal cells via particle bombardment. Confocal microscopy analysis indicated that the OsRMC-Citrine fusion protein was localized not only in the plasma membrane region but also in the extracellular cell wall region of onion epidermal cells after plasmolysis (Fig. 4). This result confirms that OsRMC, as a member of the secretory DUF26 proteins with only an extracellular domain and without a transmembrane domain or a kinase domain, is a secreted apoplastic protein.
The sequences of various cis-acting elements, including ABREs, DREs, LTREs, MYBRS, and MYCRS, were predicted in the 1,777-bp promoter region of OsRMC (Supplemental Fig. S3D). These cis elements and their respective transcription factors have important roles in abiotic stress responses (Nakashima and Yamaguchi-Shinozaki, 2006). We performed real-time Q-PCR, relative GUS activity analysis of OsRMC promoter∷GUS transgenic rice plants, and western blotting and detected the up-regulated expression profile of OsRMC in response to salt stress (Fig. 5, B–D). These results suggested that OsRMC has some functions in the response to salt stress.
In the salt stress experiments with transgenic plants, knocking down OsRMC led to growth inhibition upon 50, 100, 150, and 175 mm NaCl treatment for 10 d (Fig. 8, A–D). Although the knockdown lines show higher germination rates than the wild-type control in 175 mm NaCl treatment, in the germinated seedlings there was no apparent difference in growth reduction between the knockdown plants and control plants, both seedlings showing severe growth inhibition in such salt conditions. This result is consistent with the previous work of Jiang et al. (2007), in which knocking down OsRMC enhanced the response of transgenic rice plants to JA treatments and led to the growth inhibition of shoots and root coiling. Growth inhibition is commonly observed in transgenic plants overexpressing stress-related transcription factors that have been reported to improve tolerance to stress conditions (Ciftci-Yilmaz et al., 2007; Nakashima et al., 2007). In the plants under stress conditions, the transcript levels of certain photosynthesis-related genes and genes for carbohydrate metabolism have been found to be reduced (Sakamoto et al., 2004). Down-regulated expression of these proteins may lead to a suitable energy balance for plants under stress conditions. As a result, plant growth is suppressed and stress tolerance is increased. In the salt tolerance test assay, knockdown lines showed improved salt tolerance, as manifested in higher seed germination rates (Fig. 7, A–C), and higher seedling survival rates (Fig. 9, A and B) than the wild type.
Under salt stress conditions, the MDA contents in leaves of transgenic rice seedlings were lower than in the wild type, which indicates that the degree of lipid peroxidation was significantly reduced in transgenic plants when treated with salt stress (Fig. 9C). Meanwhile, the rice salt response genes OsDREB2A and Rab16A were up-regulated in the OsRMC knockdown transgenic plants (Fig. 9D). Expression of OsDREB2A was induced by dehydration and high-salt stresses (Dubouzet et al., 2003). Overexpression of constitutively active Arabidopsis DREB2A or maize (Zea mays) ZmDREB2A resulted in enhanced tolerance to drought stress in transgenic Arabidopsis plants (Sakuma et al., 2006; Qin et al., 2007). Rab16A belongs to the group 2 lea gene family, which was distinctly induced by salt and abscisic acid treatment (Mundy and Chua, 1988). Transgenic tobacco that overexpressed Rab16A displayed increased salt tolerance (RoyChoudhury et al., 2007). These results indicate that OsRMC negatively regulates the salt tolerance of rice plants and may be involved in the injury response of plants (Zhu, 2002; Koornneef and Pieterse, 2008).
Interestingly, it was found that the OsRMC protein was also regulated by pathogen inoculation (Kim et al., 2003) and the wound response (Shen et al., 2003), except for JA and salt stress in rice. Similarly, other DUF26 receptor protein kinases have been reported to be involved in pathogen infection, wound responses, and responses to reactive oxygen species and SA in Arabidopsis (Chen et al., 2003, 2004; Acharya et al., 2007) and rice (Nakashima et al., 2007). Therefore, the DUF26 protein family may play a broad-spectrum role in regulating the perception of injury caused by biotic and abiotic stress signals.
MATERIALS AND METHODS
Plant Material and Salt Stress Treatments
Rice (Oryza sativa subsp. japonica ‘Nipponbare’) seedlings were grown in the greenhouse at 28°C/25°C, 16 h of light/8 h of dark, and 50% humidity (Kawasaki et al., 2001). Ten-day-old rice plants were used for salt stress treatments by 200 mm NaCl solution and treated for 1, 3, and 6 h. Roots were harvested immediately for apoplastic protein extraction. For germination assays, seeds were soaked in NaCl solution and scored for germination based on whether or not the shoot length exceeded half of the seed length. For growth assay, plants were grown in 1/2MS solid medium containing NaCl for 10 d and used for growth measurements (Hu et al., 2006). For survival experiments, 10-d-old rice plants were treated with NaCl solution for 4 d and allowed to recover for 1 week. Seedlings that could not grow were considered dead (Xiong and Yang, 2003). All of these salt tolerance experiments were repeated at least three times.
Rice Transformation
Rice seeds were sterilized and cultured on Murashige and Skoog plus (4 mg L−1 2,4-dichlorophenoxyacetic acid) medium for 4 weeks in the dark at 28°C to induce embryogenic callus for Agrobacterium tumefaciens-mediated transformation, as described by Yang et al. (2004). The positively transformed calli were selected by hygromycin B (50 mg L−1) and differentiated on differentiation medium (0.5 mg L−1 naphthylacetic acid and 3 mg L−1 6-benzyladenine). The positive calli that generated roots and shoots were transferred to 1/2MS medium to develop into T0 seedlings.
Apoplast and Total Soluble Protein Extraction
Roots were cut into approximately 5-cm segments and washed with deionized water as rapidly as possible. Segments were vacuum infiltrated for 15 min in deionized water, producing a reduced pressure of 80 kPa. The vacuum was gradually released for 5 min. Segments were then dried and carefully arranged in a bundle in a Centriplus concentrator tube (Amicon). The tubes were centrifuged at 900g for 15 min at 4°C to extract the apoplast solution. The apoplast solution was lyophilized and stored at −80°C before analysis.
After the apoplastic solution had been extracted, the root tissue was ground to powder and extracted with 50 mm Tris-HCl buffer plus protease inhibitors, stirred for 15 min at 4°C, and centrifuged at 25,000g for 30 min at 4°C. The supernatant was used as total soluble protein for further analysis.
Testing Apoplastic Extract for Cytoplasmic Contamination
MDH activity was assayed by a modified method from Husted and Schjoerring (1995) in a reaction mixture containing 0.17 mm oxaloacetic acid, 0.094 mm β-NADH disodium salt, and 0.1 m Tris buffer, pH 7.4. Enzyme activity was assayed using a spectrophotometer at 340 nm by adding a 100-μL extract to 3 mL of assay medium at 25°C.
For immunoblots against tubulin, apoplastic proteins and total soluble proteins were separated by 12.5% SDS-PAGE and stained with Coomassie Brilliant Blue R-250 or transferred to a polyvinylidene difluoride membrane. The membrane was probed with anti-α-tubulin monoclonal antibody (Sigma, 1:2,000 dilution). Alkaline phosphatase-conjugated secondary antibody (Sigma, 1:10,000) was used to develop the blot.
2-DE
After quantification, 250 μL of buffer containing 150 μg of protein was loaded onto 13-cm isoelectric focusing strips, pH 3 to 10 linear gradient (Amersham Biosciences). 2-DE was carried out as described (Blum et al., 1987; Gorg et al., 1998) with minor modifications. Isoelectric focusing was conducted at 20°C using an IPGphore (Amersham Biosciences). The running conditions were as follows: 30 V for 12 h, 500 V for 1 h, 1,000 V for 1 h, and finally 8,000 V for 2 h. For the second dimension, the strips were incubated in an equilibration buffer containing 15 mm dithiothreitol for 20 min as the first step and then replaced by 2.5% iodoacetamide as the second step. The strips were placed on top of 12.5% polyacrylamide gels for SDS-PAGE according to Laemmli (1970) and sealed with 0.5% agarose. The electrophoresis was carried out at 10 mA per gel for 20 min and then 20 mA per gel by Hoefer SE 600 (Amersham Biosciences). The gels were stained with Coomassie Brilliant Blue R-250.
Image Analysis
The stained gels were scanned using Labscan software with image scanner (Amersham Biosciences). All experiments were repeated at least three times. Then, the gels were analyzed using Image Master 2D Elite software version 4.01 (Amersham Biosciences). After spot detection and normalization (in the total spot volume mode), the protein spots were matched and their volume or abundance was determined. A criterion of P < 0.05 was used to define significant differences when analyzing parallel spots between groups with one-way ANOVA using GraphPad Prim4 (GraphPad Software).
Protein Identification Using MALDI-TOF MS
In-gel digestion was performed according to Fulda et al. (2000). The mass spectra were recorded using a MALDI-TOF MS system (Bruker Autoflex). All obtained peptide mass fingerprints were searched against the National Center for Biotechnology Information database using Mascot (http://www.matrixscience.com) with a mass accuracy of 50 ppm in the parent ion mass. To denote a protein as an unambiguous identification, the following criteria were used: coverage of the mature protein by the matching peptides must reach a minimum of 15%, and at least five independent peptides should match within the protein sequence.
Molecular Cloning and Vector Construction
To construct 35S∷OsRMC-YFP transient expression vector, the OsRMC 774-bp coding sequence containing a full-length ORF without a stop codon was amplified by reverse transcription (RT)-PCR, digested by NcoI, and ligated to the pAVA321 vector (von Arnim et al., 1998). To construct OsRMCpro∷GUS vector, the 1,777-bp DNA sequence from upstream of the OsRMC ATG (start codon) was amplified, digested by HindIII and XbaI, and ligated to the pCAMBIA1300 vector. To construct the RNAi vector, the 428-bp coding sequence was amplified, digested by SacI and SpeI and then BamHI and KpnI, and ligated to the pTCK303 vector (Wang et al., 2004). To construct Ubi-1pro∷OsRMC-GUS overexpression vector, the 774-bp coding sequence was amplified, digested by BamHI, and ligated to the pCAMBIA1300 vector. All primer sequences are listed in Supplemental Table S2.
Histochemical Analysis of OsRMC Promoter∷GUS Activity
Histochemical analysis of GUS activity was according to Jefferson et al. (1987). Samples from different tissues of OsRMCpro∷GUS T1 transgenic lines were vacuum infiltrated for 15 min in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid buffer and then incubated at 37°C for 12 h. After staining, the organs were destained in 70% ethanol several times until the chlorophyll was removed. GUS-positive samples were examined with a stereoscopic zoom microscope (Nikon), and digital images were recorded. Quantitative analysis of GUS activity was performed as described by Nakashima and Yamaguchi-Shinozaki (2002). The results represent averages from three independent experiments.
Subcellular Localization of OsRMC-Citrine Fusion Protein
The plasmids of the OsRMC-Citrine fusion protein construct and Citrine empty vector, which was used as a control, were transformed into the onion (Allium cepa) epidermal cells by particle bombardment in the Bio-Rad PDS-1000/He system according to the protocol. The transformed epidermal cells were cultured on Murashige and Skoog medium for 22 h and then treated with 0.9 m mannitol for plasmolysis. Observation was performed with a confocal microscope (Zeiss 510), and digital images were recorded.
Real-Time Q-PCR
The shoots of 10-d-old rice seedlings were collected for total RNA extraction (Trizol reagent; Invitrogen). Five hundred nanograms of total RNA was used as template for the RT reaction to synthesize the first-strand cDNA, which was used for SYBR Green-monitored real-time Q-PCR as described in the protocol. The analysis was performed with the use of an ABI PRISM 7000 real-time PCR system (Applied Biosystems).
Southern Blot
The shoots of 10-d-old rice seedlings were collected for genomic DNA isolation and purification as described by Sambrook et al. (1989). Digested by HindIII and EcoRI, the fractioned DNA was electrophoresed on a 0.8% agarose gel at 60 V for 6 h and denaturized for blotting. The Hygromycin B-resistant gene from the vectors pCAMBIA1300 and pTCK303 was amplified and labeled with [α-32P]dCTP as a probe for hybridization. The image was scanned by Typhoon 9200.
Protein Purification and Antibody Preparation
The OsRMC coding sequence containing a full-length ORF without a stop codon was amplified by RT-PCR, digested by BamHI and EcoRI, and ligated to the pGEX-2T vector. Transformed into Escherichia coli strain BL21, the recombinant proteins extracted from the bacteria were purified on a GSTrap FF column according to the manufacturer's (Amersham Biosciences) protocol. The antibody of OsRMC-GST was prepared by the Tailun Biological Technology company and tested by western blot.
Determination of MDA Content
The leaves of 10-d-old rice seedlings were weighed and homogenized in 1 mL of 10% TCA solution. The homogenate was centrifuged, and the supernatant was added to 0.6% thiobarbituric acid in 10% TCA. The mixture was incubated in boiling water for 15 min, and the reaction was stopped in an ice bath. Then, the samples were centrifuged and the absorbance of the supernatant was measured at 450, 532, and 600 nm. MDA contents (nmol g−1 fresh weight) were calculated by the following formula: [6.45(A532 − A600) − 0.56A450]/fresh weight.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAL87185.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MALDI-TOF mass fingerprint of OsRMC.
Supplemental Figure S2. LC-MS/MS results showing that spot 1 and spot 6 are identical proteins.
Supplemental Figure S3. Bioinformatic analysis for OsRMC.
Supplemental Table S1. Salt stress-regulated rice root apoplastic proteins.
Supplemental Table S2. Primer sequences used in gene construction and real-time Q-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Kang Chong from the Institute of Botany, Chinese Academy of Sciences, for providing the pTCK303 vector.
This work was supported by the Chinese Key National Basic Research and Development Program (grant nos. 2006CB100100 and 2006CB910600), the National Science Foundation of China (grant nos. 30600043 and 30870200), and the Natural Science Foundation of Hebei Province in China (grant nos. C2006000142 and C2008000171).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yi Guo (guoyi@mail.hebtu.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abbasi FM, Komatsu S (2004) A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4 2072–2081 [DOI] [PubMed] [Google Scholar]
- Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R (2007) Overexpression of CRK13 an Arabidopsis cysteine-rich receptor-like kinase results in enhanced resistance to Pseudomonas syringae. Plant J 50 488–499 [DOI] [PubMed] [Google Scholar]
- Besse I, Buchanan BB (1997) Thioredoxin-linked plant and animal processes: the new generation. Bot Bull Acad Sin 38 1–11 [Google Scholar]
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8 93–99 [Google Scholar]
- Bolte S, Schiene K, Dietz KJ (2000) Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol Biol 42 923–935 [DOI] [PubMed] [Google Scholar]
- Brune A, Urbach W, Dietz KJ (1994) Compartmentation and transport of zinc in barley primary leaves as basic mechanisms involved in zinc tolerance. Plant Cell Environ 17 153–162 [Google Scholar]
- Chen K, Du L, Chen Z (2003) Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol 53 61–74 [DOI] [PubMed] [Google Scholar]
- Chen K, Fan B, Du L, Chen Z (2004) Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol 56 271–283 [DOI] [PubMed] [Google Scholar]
- Chen Z (2001) A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol 126 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282 9260–9268 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585 [DOI] [PubMed] [Google Scholar]
- Dani V, Simon WJ, Duranti M, Croy RRD (2005) Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 5 737–745 [DOI] [PubMed] [Google Scholar]
- del Carmen Córdoba-Pedregosa M, Córdoba F, Villalba JM, González-Reyes JA (2003) Zonal changes in ascorbate and hydrogen peroxide contents, peroxidase, and ascorbate-related enzyme activities in onion roots. Plant Physiol 131 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ (1997) Functions and responses of the leaf apoplast under stress. Prog Bot 58 221–254 [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33 751–763 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fuhrer J (1982) Ethylene biosynthesis and cadmium toxicity in leaf tissue of beans (Phaseolus vulgaris L.). Plant Physiol 70: 162–167 [DOI] [PMC free article] [PubMed]
- Fulda S, Huang F, Nilsson F, Hagemann M, Norling B (2000) Proteomics of Synechocystis sp strain PCC 6803: identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur J Biochem 267 5900–5907 [DOI] [PubMed] [Google Scholar]
- Gorg A, Boguth G, Obermaier C, Weiss W (1998) Two-dimensional electrophoresis of proteins in an immobilized pH 4∼12 gradient. Electrophoresis 19 1516–1519 [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J Biol Chem 276 29188–29194 [DOI] [PubMed] [Google Scholar]
- Haffani YZ, Silva NF, Goring DR (2004) Receptor kinase signaling in plant. Can J Bot 82 1–15 [Google Scholar]
- Heath RL, Parker L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125 189–198 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 35 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted S, Schjoerring JK (1995) Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol 109 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R (2008) Recent advances in plant cell wall proteomics. Proteomics 8 893–908 [DOI] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF (2006) Cell wall proteins: a new insight through proteomics. Trends Plant Sci 11 33–39 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Li J, Xu Y, Han Y, Bai Y, Zhou G, Lou Y, Xu Z, Chong K (2007) RNAi knockdown of Oryza sativa root meander curling led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ 30 690–699 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Cho KS, Yu S, Kim SG, Hong JC, Han CD, Bae DW, Nam MH, Kang KY (2003) Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics 3 2368–2378 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiol 146 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lease KA, Walker JC (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Muehling KH (2001) Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111 457–465 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35 D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y (1996) Phytosulfokine sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA 93 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk M, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G (2000) Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12 165–178 [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil JC, van der Hoorn RAL (2008) Enzyme-inhibitor interactions at the plant-pathogen interface. Curr Opin Plant Biol 11 1–9 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Yoshida R, Shinozaki K (2000) MAP kinase cascades in Arabidopsis: their roles in stress and hormone responses. Results Probl Cell Differ 27 29–38 [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17 646–653 [DOI] [PubMed] [Google Scholar]
- Mundy J, Chua NH (1988) Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J 7 2279–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Ligterink W, Meskiene I, Calderini O, Beyerly J, Musgrave A, Hirt H (1999) Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J 20 381–388 [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer HJ, Ter Riet B, Hirt H, Frank W, Bartels D, Musgrave A (2000) Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J 22 147–154 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51 617–630 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K (2002) Use of β-glucuronidase to show dehydration and high-salt gene expression. In JF Jackson, HF Linsken, RB Inman, eds, Molecular Methods of Plant Analysis, Vol 22. Testing for Genetic Manipulation in Plants. Springer-Verlag, Berlin, pp 37–61
- Nakashima K, Yamaguchi-Shinozaki K (2006) Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plant 126 62–71 [Google Scholar]
- Ndimba BK, Chivasa S, Hamilton JM, Simon WJ, Slabas AR (2003) Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3 1047–1059 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10 1–6 [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Schjoerring JK (1998) Regulation of apoplastic NH4+ concentration in leaves of oilseed rape. Plant Physiol 118 1361–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253 895–898 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50 54–69 [DOI] [PubMed] [Google Scholar]
- Ramanjulu S, Kaiser W, Dietz KJ (1999) Salt and drought stress differentially affect the accumulation of extracellular proteins in barley. Z Naturforsch 54 337–347 [Google Scholar]
- RoyChoudhury A, Roy C, Sengupta DN (2007) Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep 26 1839–1859 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2 1131–1145 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schopfer CR, Nasrallah ME, Nasrallah JB (1999) The male determinant of self-incompatibility in Brassica. Science 286 1697–1700 [DOI] [PubMed] [Google Scholar]
- Shen S, Jing Y, Kuang T (2003) Proteomics approach to identify wound-response related proteins from rice leaf sheath. Proteomics 3 527–535 [DOI] [PubMed] [Google Scholar]
- Swapan KD, Muthukrishnan S (1999) Pathogenesis Related Proteins in Plants. CRC Press, New York, pp 1–278
- Takeda H, Kotake T, Nakagawa N, Sakurai N, Nevins DJ (2003) Expression and function of cell wall-bound cationic peroxidase in asparagus somatic embryogenesis. Plant Physiol 131 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel J, Grenier J, Potvin C, Asselin A (1998) Several thaumatin-like proteins bind to β-1,3-glucans. Plant Physiol 118 1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11 329–334 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221 35–43 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L). Plant Mol Biol Rep 22 409–417 [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SP, Tang ZC, Sun WA, Sun WN (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5 235–244 [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167 281–288 [Google Scholar]
- Young SA, Guo A, Guikema JA, White FF, Leach JE (1995) Rice cationic peroxidase accumulates in xylem vessels during incompatible interaction with Xanthomonas oryzae pv. oryzae. Plant Physiol 107 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.