Abstract
Transgenic tomato (Solanum lycopersicum) plants in which either mitochondrial malate dehydrogenase or fumarase was antisense inhibited have previously been characterized to exhibit altered photosynthetic metabolism. Here, we demonstrate that these manipulations also resulted in differences in root growth, with both transgenics being characterized by a dramatic reduction of root dry matter deposition and respiratory activity but opposite changes with respect to root area. A range of physiological, molecular, and biochemical experiments were carried out in order to determine whether changes in root morphology were due to altered metabolism within the root itself, alterations in the nature of the transformants' root exudation, consequences of alteration in the efficiency of photoassimilate delivery to the root, or a combination of these factors. Grafting experiments in which the transformants were reciprocally grafted to wild-type controls suggested that root length and area were determined by the aerial part of the plant but that biomass was not. Despite the transgenic roots displaying alteration in the expression of phytohormone-associated genes, evaluation of the levels of the hormones themselves revealed that, with the exception of gibberellins, they were largely unaltered. When taken together, these combined experiments suggest that root biomass and growth are retarded by root-specific alterations in metabolism and gibberellin contents. These data are discussed in the context of current models of root growth and biomass partitioning.
The structure of the plant tricarboxylic acid (TCA) cycle has been established for decades (Beevers, 1961), and in vitro studies have established regulatory properties of many of its component enzymes (Budde and Randall, 1990; Millar and Leaver, 2000; Studart-Guimarães et al., 2005). That said, relatively little is known, as yet, regarding how this important pathway is regulated in vivo (Fernie et al., 2004a; Sweetlove et al., 2007). Indeed, even fundamental questions concerning the degree to which this pathway operates in illuminated leaves (Tcherkez et al., 2005; Nunes-Nesi et al., 2007a) and the influence it has on organic acid levels in fruits (Burger et al., 2003) remain contentious. Furthermore, in contrast to many other pathways of primary metabolism, the TCA cycle has been subjected to relatively few molecular physiological studies. To date, the functions of pyruvate dehydrogenase, citrate synthase, aconitase, isocitrate dehydrogenase, succinyl-CoA ligase, fumarase, and malate dehydrogenase have been studied via this approach (Landschütze et al., 1995; Carrari et al., 2003; Yui et al., 2003; Nunes-Nesi et al., 2005, 2007a; Lemaitre et al., 2007; Studart-Guimarães et al., 2007); however, several of these studies were relatively cursory. Despite this fact, they generally corroborate one another, with at least two studies providing clear evidence for an important role of the TCA cycle in flower development (Landschütze et al., 1995; Yui et al., 2003) or in the coordination of photosynthetic and respiratory metabolisms of the illuminated leaf (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007a).
In our own studies on tomato (Solanum lycopersicum), we have observed that modulation of fumarase and mitochondrial malate dehydrogenase activities leads to contrasting shoot phenotypes, with the former displaying stunted growth while the later exhibited an enhanced photosynthetic performance (Nunes-Nesi et al., 2005, 2007a). We were able to demonstrate that the stunted-growth phenotype observed in aerial parts of the fumarase plants was a consequence of altered stomatal function (Nunes-Nesi et al., 2007a), whereas the increased photosynthetic performance of the mitochondrial malate dehydrogenase seems likely to be mediated by the alterations in ascorbate metabolism exhibited by these plants (Nunes-Nesi et al., 2005; Urbanczyk-Wochniak et al., 2006). In keeping with the altered rates of photosynthesis in these antisense plants, the fruit yield of fumarase and mitochondrial malate dehydrogenase plants was decreased and increased, respectively. However, the root biomass of both transgenics was significantly reduced (Nunes-Nesi et al., 2005, 2007a). These observations were somewhat surprising given that it is estimated that 30% to 60% of net photosynthate is transported to root organs (Merckx et al., 1986; Nguyen et al., 1999; Singer et al., 2003). When taken together, these results suggest that the root phenotype must result from either an impairment of translocation or a root-specific effect. Neither of these explanations is without precedence, with inhibition of the expression of Suc transporters (Riesmeier et al., 1993; Gottwald et al., 2000) resulting in dramatically impaired root growth while organic acid exudation itself has been implicated in a wide range of root organ functions, including nutrient acquisition (de la Fuente et al., 1997; Imas et al., 1997; Neumann and Römheld, 1999; López-Bucio et al., 2000; Anoop et al., 2003; Delhaize et al., 2004), metal sequestration (Gillooly et al., 1983; de la Fuente et al., 1997; Cramer and Titus, 2001), and microbial proliferation in the rhizosphere (Lugtenberg et al., 1999; Weisskopf et al., 2005). In addition to the putative mechanisms listed above, the TCA cycle could be anticipated to play a vital role in meeting the high energy demands of nitrogen fixation and polymer biosynthesis associated with rapidly growing heterotrophic organs (Pradet and Raymond, 1983; Dieuaide-Noubhani et al., 1997; Stasolla et al., 2003; Deuschle et al., 2006). In keeping with this theory, alteration of the energy status of roots and other heterotrophic tissue has been documented to positively correlate with elevated biomass production (Anekonda, 2001; Regierer et al., 2002; Carrari et al., 2003; Lovas et al., 2003; Geigenberger et al., 2005). Here, we performed a detailed physiological, molecular, and biochemical evaluation of whole plant and root metabolism of the mitochondrial malate dehydrogenase and fumarate antisense tomato lines. In this manner, we broadly assessed biochemical changes in the root, including the levels of several major phytohormones, as well as dissected which characteristics were influenced by aerial parts of the plant. The results obtained are discussed both with respect to the regulation of the TCA cycle per se and within the context of the determination of root morphology and growth.
RESULTS
Establishment of the Experimental System
Given the previous observations that plants exhibiting reduced activities of several enzymes of the mitochondrial TCA cycle display a restricted root biomass (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007a), we set out to better understand the mechanistic basis for this phenomenon. For this purpose, we decided to focus on well-characterized transgenic lines in which the expression of fumarase (FL lines) or the mitochondrial malate dehydrogenase (mMDH lines) was repressed. Three potential explanations could be postulated to explain the observed alterations in root morphology. The root phenotype could arise from (1) altered photoassimilate supply from source organs, (2) root-specific effects, (3) an altered root exudation profile, or a combination of two or more of these factors. The first step in this study was to find suitable experimental growth conditions to assess the contributions of these factors. Comparing endometabolite contents between soil-grown and vermiculite-grown 5-week-old tomato roots provided a practical incentive to grow the plants on vermiculite (data not shown). In accordance with current literature (Bertin et al., 2003), the apical regions of the mature root were found to be characterized by high organic acid content. For these reasons, subsequent studies were focused on this tissue.
Diurnal Variation in Root Endometabolite and Exometabolite Profiles
Having established the experimental system with which to address our questions, we next turned our attention to the preliminary characterization of root metabolite exudation. As a first experiment, we analyzed the levels of both endogenous root tip (the entire 100-mm region proximal to the apex) metabolites and exuded metabolites during a 24-h cycle. Very few clear trends were apparent in the data from the endogenous metabolites. All amino acids analyzed remained stable throughout the time course (Supplemental Fig. S1A), and this was also true for the majority of the organic acid and sugar compounds (Supplemental Fig. S1, B and C). Tomato root systems subjected to an additional 2 d of darkness displayed a sharp decrease in both organic acid and sugar (mainly Suc and Fru) contents (Supplemental Fig. S1, E and F). By contrast, the amino acid levels seemed either to remain unaltered by this experimental treatment (e.g. Ser, Met, Ala, and Asp) or peaked during defined times in the extended night period (e.g. Ile and Glu; Supplemental Fig. S1D).
In the case of the exometabolites, a more pronounced circadian-like rhythm could be observed (Supplemental Fig. S2). The pattern of organic acid displayed a peak toward the end of the light period, with the lowest reported values for organic acids and sugars in the morning and during midday (Supplemental Fig. S2, B and C). As was observed for the endometabolites, there were few trends in the levels of amino acids, although Lys and Cys both peaked toward the end of the light period (Supplemental Fig. S2A). Most exometabolites displayed similar rhythmic fluctuations on the extension of the night. However, the levels of Glu, isocitrate, citrate, Suc, Glc, and Fru were dramatically decreased on extended darkness, and their patterns displayed a dramatic decrease in amplitude (Supplemental Fig. S2, D–F; Supplemental Table S1).
Confirmation of Inhibition of Malate Dehydrogenase and Fumarase Activities, and Consequently of Carbon Dioxide Evolution, in Roots of the Transgenic Lines
As a first experiment, the activities of malate dehydrogenase and fumarase were detected in root extracts of the previously characterized transgenic lines (Nunes-Nesi et al., 2005, 2007a). These measurements demonstrated that both sets of transgenics were characterized by a dramatic decrease in the root activities of their target enzymes (Table I). The FL lines were characterized by decreases of root activity down to 18% of that observed in the wild type, whereas the mMDH lines were characterized by decreases of total malate dehydrogenase activity down to 39% of that observed in the wild type (Table I). When these relative values are compared with the previously recorded changes in leaf activity, it appears that the mitochondrial malate dehydrogenase accounts for a higher proportion of the total cellular activity in roots than it does in leaves (Nunes-Nesi et al., 2005). By contrast, the inhibition of fumarase activity in the roots is marginally lower than that observed in leaves (Nunes-Nesi et al., 2007a). In keeping with this, quantification of the protein abundance in the transgenics revealed that it was in accordance with the degree of reduction in fumarase activity (data not shown). Furthermore, 14CO2 evolution following the incubation of root material in [U-14C]Glc indicated that the transgenic lines displayed only 20% to 30% of the CO2 release of the wild type, implying a dramatic impairment of TCA cycle activity in roots of the transformants (data not shown). This respiratory restriction was qualitatively consistent with those previously observed in other tissues, although notably of much greater magnitude in roots (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007a).
Table I.
Characterization of enzyme activities in roots of 5-week-old FL and mMDH antisense transgenic plants
Values are presented as means ± se of six individual plants per line; values shown in boldface were determined by the t test to be significantly different (P < 0.05) from the wild type. –, Not determined.
| Enzyme Activities | Wild Type | mMDH7 | mMDH8 | mMDH21 | FL11 | FL41 | FL63 |
|---|---|---|---|---|---|---|---|
| μmol g−1 fresh weight min−1 | |||||||
| Total MDH-NAD dependent | 3,420 ± 310 | 1,540 ± 140 | 1,330 ± 140 | 1,780 ± 160 | – | – | – |
| Fumarase | 40.2 ± 5.1 | – | – | – | 8.72 ± 0.9 | 7.12 ± 0.9 | 22.5 ± 2.1 |
Root Morphology and Physiology in the Mitochondrial Malate Dehydrogenase and Fumarase Transformants
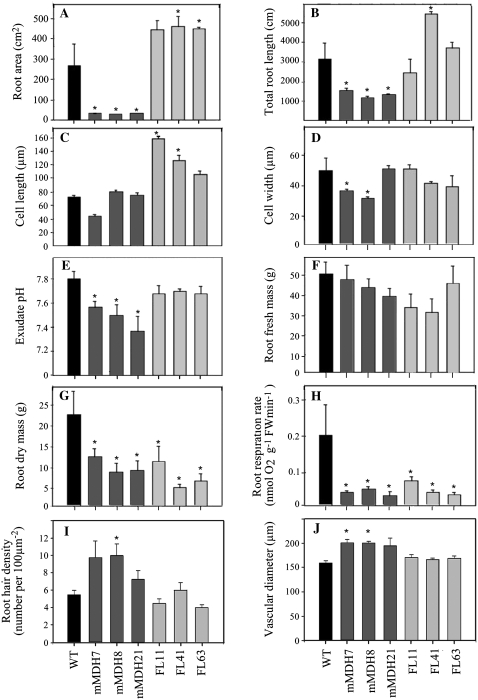
When grown in vermiculite, the transgenic lines displayed dramatic alterations in root area that were markedly reduced in the mMDH lines and significantly enhanced in two of the three FL lines (Fig. 1A). A similar picture was apparent in root length, although in this instance the increase was only significant in one of the FL lines (line FL41; Fig. 1B). Measuring the cell length of equivalent cells from the root tip revealed that both lines FL11 and FL41 were significantly longer (up to 2-fold as long) but that the other lines were invariant in this parameter (Fig. 1C). Perhaps surprisingly, the FL lines exhibited an invariant cell width whereas lines mMDH7 and mMDH8 were characterized as having reduced cell widths (Fig. 1D). When the exudates of the transgenics were harvested (as described in “Materials and Methods”) and their pH quantified, it became apparent that the exudates from all three mMDH lines were considerably more acidic than those of the wild type (Fig. 1E). Intriguingly, there was only a minor, and nonsignificant, decrease in total root fresh weight in both sets of transformants (Fig. 1F) but relatively dramatic reductions in root dry mass (down to 25% of that observed in the wild type; Fig. 1G). Direct analysis of the rate of root respiration, however, confirmed that this was impaired in both sets of transgenics, being reduced to levels corresponding to approximately 20% of those observed for the wild type (Fig. 1H). The root hair density was largely unaltered in the transgenic lines, being increased in line mMDH8 but invariant in all other lines (Fig. 1I). Similarly, mMDH7 and mMDH8 displayed an increased vascular diameter, but all other lines were invariant from the wild type (Fig. 1J). Furthermore, both the number of secondary roots and the number of cell layers were invariant in all transgenic lines in comparison with the wild type (data not shown). Thus, despite displaying similar reductions in root respiration and dry mass, the transformants displayed markedly different root phenotypes, with the mitochondrial malate dehydrogenase plants exhibiting stunted root growth and reduced root area while roots of the FL lines were, if anything, longer and had a greater surface area than those of the wild type.
Figure 1.
Root morphologies of transgenic tomato roots down-regulated in mMDH or fumarase activity after 5 weeks of growth. Root area measurements (A), total root length (B), cell length (C), and cell diameter (D) are shown. In addition, the root exudate pH (E), root fresh mass (F), root dry mass (G), root respiration measurements (H), root hair density (I), and vascular diameter (J) were determined. The values are means ± se of six individual plants per line. Asterisks indicate values determined by the t test to be significantly different from the wild type (WT; P < 0.05).
Root Carbohydrate, Adenylate, and Redox Contents of the Transformants
We next decided to evaluate the content of carbohydrates in the roots of material harvested at the end of the light period from 5-week-old vermiculite-grown plants. Soluble sugars and starch were extracted and measured using established spectrophotometric methods (Müller-Röber et al., 1992; Table II). The carbohydrate content of wild-type roots was similar to that previously reported (Carrari et al., 2005; Lou et al., 2007). These studies revealed that the FL lines were characterized by decreased levels of Glc (significantly so in lines FL11 and FL63), Fru (significantly so in lines FL11 and FL41), and Suc (significantly only in line FL63). By contrast, the mMDH lines were invariant in all of these parameters with respect to the wild type. However, it is perhaps important to note that neither the total soluble carbohydrate content nor the protein content (Table II) of the FL lines was significantly different from those of the wild type, with the levels of starch being decreased in both sets of transformants (significantly so in lines FL41 and all mMDH lines; Table II).
Table II.
Effects of decreased fumarase and mMDH activity on sugars, starch, and protein contents of 5-week-old mature antisense root tips
Data are presented as means ± se of six individual plants per line. Values in boldface were determined by the t test to be significantly different (P < 0.05) from the wild type. Values are presented as μmol g−1 fresh weight, except for protein, which is μg g−1 fresh weight.
| Sugars/Protein | Wild Type | mMDH7 | mMDH8 | mMDH21 | FL11 | FL41 | FL63 |
|---|---|---|---|---|---|---|---|
| Glc | 48.6 ± 6.7 | 35.2 ± 5.6 | 48.9 ± 4.7 | 33.1 ± 2.5 | 31.0 ± 2.1 | 35.0 ± 5.9 | 29.5 ± 4.1 |
| Fru | 38.1 ± 0.5 | 43.4 ± 2.7 | 40.6 ± 5.9 | 36.9 ± 7.4 | 29.0 ± 2.7 | 34.2 ± 1.0 | 24.3 ± 5.2 |
| Suc | 11.6 ± 1.9 | 15.0 ± 2.3 | 14.2 ± 2.1 | 12.7 ± 1.0 | 11.3 ± 1.2 | 9.9 ± 1.6 | 5.5 ± 1.1 |
| Total soluble sugars (hexose equivalents) | 89.0 ± 18.4 | 90.5 ± 6.0 | 104.9 ± 7.4 | 72.1 ± 5.7 | 72.7 ± 4.3 | 74.2 ± 5.8 | 54.0 ± 5.5 |
| Starch | 15.1 ± 2.7 | 6.7 ± 1.6 | 8.2 ± 0.7 | 4.7 ± 0.7 | 8.8 ± 1.1 | 9.1 ± 0.9 | 12.6 ± 1.7 |
| Protein | 147 ± 15 | 196 ± 54 | 152 ± 53 | 129 ± 13 | 198 ± 53 | 124 ± 6 | 187 ± 21 |
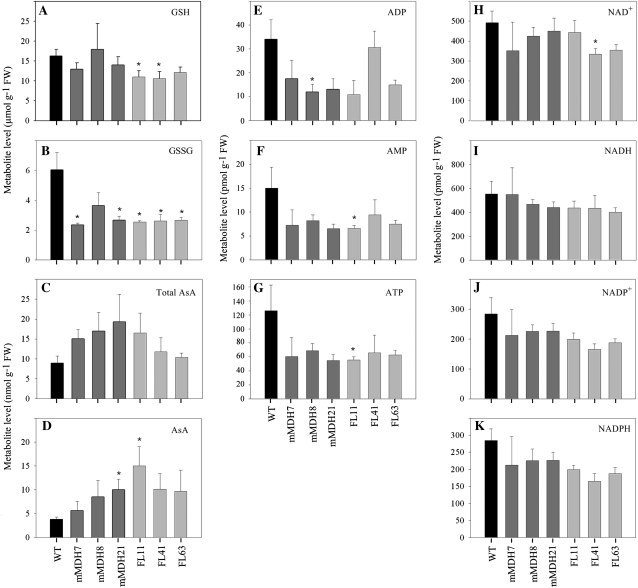
Redox profiling of the transgenic lines revealed a significant decreases in reduced glutathione in the FL lines, while oxidized glutathione levels were reduced to approximately 60% of that found in the wild type in both the FL and mMDH lines (Fig. 2). In addition, a tendency of increase in total ascorbate was observed in both the mMDH and FL lines, although in both sets of plants ascorbate levels were only significantly elevated in a single line (mMDH21 and FL11, respectively). Furthermore, the deduced ratio of total ascorbate to ascorbate was largely unaltered in the mMDH lines but dramatically lower in the FL lines. Despite these changes, the levels of reducing equivalents were essentially invariant in the transgenic lines, with only a single significant difference being observed (the mild decrease in NAD content in line FL41). In contrast, the levels of adenylates appeared to be somewhat reduced in the lines, significantly so in the case of line mMDH8 (ADP) and line FL11 (AMP and ATP), but the ATP-ADP ratios and the adenylate energy charges of the transformants were invariant from the wild type (Fig. 2).
Figure 2.
Redox levels in 5-week-old-plants of reduced glutathione (GSH; A), oxidized glutathione (GSSG; B), total ascorbate (Total AsA; C), ascorbate (AsA; D), adenylate (ADP, AMP, ATP; E–G), and reducing equivalents (NAD+, NADH, NADP+, NADPH; H–K) in the root tissue of down-regulated mMDH and FL transgenic lines. The values are means ± se of six individual plants. Asterisks indicate values determined by the t test to be significantly different from the wild-type (WT) control (P < 0.05). FW, Fresh weight.
Analysis of Endometabolites in the Transformants
To further characterize the metabolic changes in the roots of these lines, an established gas chromatography-mass spectrometry (GC-MS)-based metabolite-profiling method (Fernie et al., 2004b) was employed. These analyses allowed the detection of the relative levels of over 50 metabolites. The mMDH lines displayed relatively few significant alterations in the levels of these metabolites (Table III). That said, clear trends toward increases in a number of metabolites were noticeable, including those in Ara (significant in lines mMDH7 and mMDH8), Xyl, maltose, Tyr, tyramine, Trp (significant in line mMDH7 only), and succinate (significant in lines mMDH8 and mMDH21). In addition, 3-phosphoglycerate decreased significantly in lines mMDH8 and mMDH21. By contrast, the FL lines were characterized by greater and more consistent changes, with most of the metabolites that were altered in the transgenics exhibiting decreased levels with respect to the wild type (Table III).
Table III.
Relative metabolite contents of roots from mMDH and FL lines
Metabolites were determined as described in “Materials and Methods.” Data are normalized with respect to mean response calculated for the wild type (to allow statistical assessment in the same way). Values are presented as means ± se of six individual plants per line, and values in boldface were determined by t test to be significantly different (P < 0.05) from the wild type.
| Metabolite | Wild Type | mMDH7 | mMDH8 | mMDH21 | FL11 | FL41 | FL63 |
|---|---|---|---|---|---|---|---|
| Aconitate | 1.00 ± 0.48 | 0.99 ± 0.35 | 0.87 ± 0.40 | 0.88 ± 0.39 | 0.56 ± 0.52 | 0.24 ± 0.35 | 0.76 ± 0.73 |
| Ala | 1.00 ± 0.30 | 1.05 ± 0.22 | 1.76 ± 0.41 | 1.22 ± 0.26 | 1.24 ± 0.26 | 2.61 ± 0.23 | 1.29 ± 0.17 |
| Ara | 1.00 ± 0.33 | 2.66 ± 0.33 | 2.91 ± 0.28 | 1.51 ± 0.39 | 1.22 ± 0.31 | 1.96 ± 0.44 | 0.83 ± 0.25 |
| Arg | 1.00 ± 0.36 | 0.92 ± 0.46 | 1.71 ± 0.49 | 0.97 ± 0.60 | 0.69 ± 0.56 | 1.54 ± 0.60 | 0.85 ± 0.60 |
| Asn | 1.00 ± 0.38 | 1.43 ± 0.30 | 1.55 ± 0.86 | 0.80 ± 0.48 | 0.53 ± 0.71 | 0.42 ± 0.60 | 0.51 ± 0.85 |
| Asp | 1.00 ± 0.47 | 0.85 ± 0.58 | 0.58 ± 0.31 | 0.41 ± 0.49 | 1.98 ± 0.50 | 0.67 ± 0.27 | 0.45 ± 0.39 |
| Citrate | 1.00 ± 0.32 | 1.43 ± 0.30 | 1.75 ± 0.31 | 0.69 ± 0.38 | 0.47 ± 0.26 | 0.23 ± 0.49 | 0.29 ± 0.48 |
| Cys | 1.00 ± 0.17 | 2.43 ± 0.44 | 2.84 ± 0.39 | 1.20 ± 0.35 | 1.31 ± 0.31 | 0.90 ± 0.25 | 0.78 ± 0.11 |
| Dehydroascorbate | 1.00 ± 0.33 | 1.25 ± 0.35 | 1.44 ± 0.53 | 0.94 ± 0.38 | 0.47 ± 0.29 | 0.64 ± 0.76 | 0.21 ± 0.24 |
| Fru | 1.00 ± 0.36 | 0.78 ± 0.55 | 1.55 ± 0.42 | 0.77 ± 0.26 | 0.17 ± 0.40 | 0.08 ± 0.53 | 0.17 ± 0.35 |
| Fru 6-P | 1.00 ± 0.47 | 1.03 ± 0.30 | 1.06 ± 0.32 | 2.03 ± 0.47 | 1.62 ± 0.33 | 0.73 ± 0.77 | 1.16 ± 0.69 |
| Fumarate | 1.00 ± 0.23 | 1.97 ± 0.58 | 2.08 ± 0.56 | 0.54 ± 0.25 | 2.19 ± 0.47 | 0.70 ± 0.30 | 0.63 ± 0.38 |
| GABA | 1.00 ± 0.21 | 1.13 ± 0.37 | 1.38 ± 0.40 | 0.85 ± 0.38 | 0.29 ± 0.22 | 0.42 ± 0.36 | 0.25 ± 0.21 |
| Gal | 1.00 ± 0.31 | 2.04 ± 0.44 | 1.25 ± 0.50 | 1.75 ± 0.57 | 0.32 ± 0.32 | 0.40 ± 0.57 | 0.20 ± 0.44 |
| Galacturonate | 1.00 ± 0.51 | 1.42 ± 0.49 | 1.87 ± 0.56 | 1.11 ± 0.49 | 1.04 ± 0.71 | 0.27 ± 0.58 | 0.87 ± 0.85 |
| Glc | 1.00 ± 0.32 | 1.27 ± 0.38 | 0.92 ± 0.48 | 1.64 ± 0.54 | 0.47 ± 0.28 | 0.26 ± 0.41 | 0.25 ± 0.40 |
| Glc 6-P | 1.00 ± 0.41 | 0.84 ± 0.22 | 1.00 ± 0.38 | 1.29 ± 0.42 | 1.50 ± 0.38 | 0.42 ± 0.76 | 1.09 ± 0.80 |
| Glu | 1.00 ± 0.41 | 0.87 ± 0.45 | 1.18 ± 0.32 | 0.97 ± 0.49 | 0.26 ± 0.42 | 0.16 ± 0.26 | 0.13 ± 0.47 |
| Gln | 1.00 ± 0.33 | 5.66 ± 0.41 | 10.49 ± 0.61 | 6.95 ± 0.54 | 0.10 ± 0.20 | 0.20 ± 0.43 | 0.06 ± 0.19 |
| Glyceric 3-P | 1.00 ± 0.25 | 0.62 ± 0.29 | 0.28 ± 0.25 | 0.35 ± 0.47 | 0.15 ± 0.31 | 0.37 ± 0.67 | 0.13 ± 0.18 |
| Glycerol 3-P | 1.00 ± 0.30 | 1.54 ± 0.25 | 1.80 ± 0.41 | 1.68 ± 0.56 | 2.36 ± 0.41 | 2.72 ± 0.22 | 0.56 ± 0.37 |
| Gly | 1.00 ± 0.13 | 2.31 ± 0.44 | 0.80 ± 0.23 | 0.70 ± 0.22 | 1.46 ± 0.18 | 0.45 ± 0.23 | 0.56 ± 0.32 |
| Isocitrate | 1.00 ± 0.27 | 1.92 ± 0.35 | 1.27 ± 0.32 | 1.17 ± 0.37 | 1.10 ± 0.49 | 0.30 ± 0.44 | 0.98 ± 0.67 |
| Ile | 1.00 ± 0.24 | 1.46 ± 0.24 | 1.55 ± 0.55 | 1.87 ± 0.52 | 1.23 ± 0.48 | 0.65 ± 0.25 | 0.83 ± 0.29 |
| l-Ascorbate | 1.00 ± 0.36 | 1.95 ± 0.53 | 1.22 ± 0.32 | 0.80 ± 0.39 | 0.64 ± 0.51 | 0.29 ± 0.33 | 0.25 ± 0.55 |
| Leu | 1.00 ± 0.31 | 1.15 ± 0.49 | 1.84 ± 0.45 | 0.72 ± 0.41 | 0.89 ± 0.46 | 0.74 ± 0.66 | 0.58 ± 0.29 |
| Lys | 1.00 ± 0.28 | 3.65 ± 0.62 | 4.26 ± 0.62 | 2.97 ± 0.39 | 1.50 ± 0.39 | 1.17 ± 0.58 | 1.20 ± 0.47 |
| Malate | 1.00 ± 0.20 | 0.94 ± 0.59 | 1.03 ± 0.27 | 0.59 ± 0.33 | 0.21 ± 0.36 | 0.13 ± 0.33 | 0.37 ± 0.32 |
| Maltose | 1.00 ± 0.36 | 2.42 ± 0.22 | 1.90 ± 0.26 | 2.15 ± 0.30 | 1.65 ± 0.27 | 0.88 ± 0.53 | 0.58 ± 0.28 |
| Mannitol | 1.00 ± 0.32 | 1.54 ± 0.34 | 1.78 ± 0.31 | 1.39 ± 0.37 | 2.13 ± 0.64 | 4.60 ± 0.90 | 8.79 ± 0.66 |
| Met | 1.00 ± 0.38 | 0.58 ± 0.34 | 0.16 ± 0.28 | 0.45 ± 0.39 | 0.15 ± 0.39 | 0.10 ± 0.48 | 0.07 ± 0.30 |
| Myoinositol | 1.00 ± 0.40 | 0.70 ± 0.30 | 0.45 ± 0.28 | 0.38 ± 0.33 | 0.27 ± 0.30 | 0.28 ± 0.48 | 0.12 ± 0.21 |
| Palmeate (16:0) | 1.00 ± 0.35 | 2.15 ± 0.38 | 1.58 ± 0.40 | 1.17 ± 0.42 | 0.88 ± 0.32 | 0.49 ± 0.27 | 0.49 ± 0.38 |
| Phe | 1.00 ± 0.30 | 1.65 ± 0.41 | 1.33 ± 0.48 | 0.85 ± 0.51 | 0.41 ± 0.48 | 0.46 ± 0.50 | 0.33 ± 0.35 |
| Pro | 1.00 ± 0.35 | 1.55 ± 0.55 | 1.11 ± 0.42 | 1.21 ± 0.65 | 1.02 ± 0.50 | 0.19 ± 0.23 | 0.81 ± 0.29 |
| Putrescine | 1.00 ± 0.30 | 2.78 ± 0.30 | 1.31 ± 0.42 | 3.24 ± 0.38 | 1.07 ± 0.27 | 1.05 ± 0.64 | 0.68 ± 0.33 |
| Quinate | 1.00 ± 0.36 | 0.78 ± 0.38 | 1.34 ± 0.30 | 1.45 ± 0.62 | 1.05 ± 0.33 | 0.60 ± 0.37 | 0.39 ± 0.27 |
| Saccharate | 1.00 ± 0.28 | 0.66 ± 0.27 | 0.61 ± 0.40 | 0.73 ± 0.30 | 0.71 ± 0.21 | 0.44 ± 0.52 | 0.17 ± 0.18 |
| Ser | 1.00 ± 0.53 | 2.20 ± 0.39 | 1.35 ± 0.49 | 0.97 ± 0.48 | 0.69 ± 0.39 | 0.64 ± 0.64 | 0.49 ± 0.21 |
| Shikimate | 1.00 ± 0.30 | 3.25 ± 0.31 | 1.01 ± 0.39 | 0.86 ± 0.24 | 0.87 ± 0.38 | 0.17 ± 0.30 | 0.25 ± 0.39 |
| Stearate (18:0) | 1.00 ± 0.33 | 6.67 ± 0.42 | 7.45 ± 0.41 | 5.15 ± 0.45 | 6.53 ± 0.27 | 1.99 ± 0.19 | 1.55 ± 0.38 |
| Succinate | 1.00 ± 0.32 | 3.68 ± 0.30 | 8.60 ± 0.31 | 8.27 ± 0.37 | 3.31 ± 0.38 | 5.74 ± 0.58 | 7.09 ± 0.24 |
| Suc | 1.00 ± 0.30 | 2.04 ± 0.44 | 1.78 ± 0.40 | 1.09 ± 0.26 | 0.92 ± 0.68 | 0.26 ± 0.36 | 0.21 ± 0.33 |
| Threonate | 1.00 ± 0.56 | 0.96 ± 0.36 | 0.81 ± 0.37 | 0.49 ± 0.35 | 0.46 ± 0.29 | 0.24 ± 0.11 | 0.31 ± 0.10 |
| Thr | 1.00 ± 0.31 | 2.45 ± 0.39 | 0.82 ± 0.20 | 1.32 ± 0.58 | 1.11 ± 0.30 | 0.71 ± 0.30 | 0.93 ± 0.35 |
| Trehalose | 1.00 ± 0.33 | 1.34 ± 0.29 | 2.25 ± 0.45 | 1.29 ± 0.36 | 1.40 ± 0.28 | 0.59 ± 0.63 | 0.58 ± 0.43 |
| Trp | 1.00 ± 0.34 | 3.13 ± 0.27 | 3.77 ± 0.53 | 1.69 ± 0.68 | 0.83 ± 0.45 | 1.27 ± 0.60 | 2.13 ± 0.50 |
| Tyramine | 1.00 ± 0.35 | 6.54 ± 0.35 | 2.76 ± 0.49 | 3.40 ± 0.50 | 1.05 ± 0.52 | 0.86 ± 0.53 | 0.56 ± 0.28 |
| Tyr | 1.00 ± 0.28 | 3.31 ± 0.35 | 5.28 ± 0.68 | 0.94 ± 0.16 | 0.91 ± 0.66 | 1.34 ± 0.67 | 1.21 ± 0.77 |
| Val | 1.00 ± 0.20 | 0.96 ± 0.23 | 1.34 ± 0.46 | 0.73 ± 0.31 | 1.05 ± 0.35 | 1.02 ± 0.56 | 0.64 ± 0.22 |
| Xyl | 1.00 ± 0.39 | 8.93 ± 0.42 | 2.08 ± 0.28 | 2.46 ± 0.33 | 1.67 ± 0.56 | 1.43 ± 0.62 | 0.58 ± 0.30 |
Analysis of Exometabolites in the Transformants
We next utilized GC-MS to determine the relative levels of metabolites excreted from the roots of transgenic and wild-type plants in order to assess the potential influence of exudation on the root phenotype. For this purpose, exudates were collected at the end of the light period as described above from all lines. As noted previously for the metabolite levels within the transgenic roots, the number and magnitude of changes observed was much greater in the FL lines than in the mMDH lines (Supplemental Table S3). The malate dehydrogenase transformants were characterized by only minor differences in their exudation profiles, namely, decreased exudation of trehalose (significant in line mMDH7) and of Glc, Gal, and Man (significant in line mMDH8). By contrast, there was a large increase in the relative levels of metabolites exuded by the roots of the FL lines (despite a relatively high level of biological variance in these samples). Assessment of the sugars and sugar derivatives revealed an increase in isomaltose, maltitol, trehalose, rhamnose, Ara, Xyl, and Man (however, this was only significant in line FL11) as well as in Glc (significant in line FL63), Gal (significant in lines FL11 and FL63), glycerol 1-phosphate (significant in line FL63), glycerol, and Rib (significant in lines FL11 and FL63). Changes were also apparent in the organic acids, with increased exudation in shikimate (significant in all lines), quinate, fumarate, and glycerate (significant in lines FL11 and FL63), and citrate, succinate, oxalate, benzoate, glutarate, malate, and saccharate (significant in line FL11). Furthermore, the levels of several amino acids were elevated in the FL line exudates, with the levels of Gly (significant in all lines), β-Ala and Asp (significant in lines FL11 and FL63), and Glu, hydoxy-Pro, Asn, Met, Thr, Glu, and Ile (significant in line FL11) all increasing. Gln levels also increased 10-fold in line FL41.
In order to evaluate the exudation data in context with that of the endometabolites, we ran a series of calibration curves of authentic standards alongside our experimental extracts to allow us to perform absolute quantification of the metabolite levels (these data can all be viewed in Supplemental Tables S2 and S3). Several interesting features arise from this data transformation. Consistent with previous reports (Carrari et al., 2005; Desbrosses et al., 2005), the absolute levels of metabolites with respect to one another are dramatically different from those found in the leaves, with a significantly higher content of organic acids found in the heterotrophic organ. Comparison of the absolute levels of metabolites within the root system with those in the exudate revealed that fumarate, shikimate, Rib, Asp, Ala, Cys, β-aminobutyrate (GABA), Gly, Ile, Phe, Thr, and Val were excreted at considerable levels relative to their cellular content (data not shown). A comparison of the relative secretion of metabolites between the transgenics and the wild type revealed a dramatic shift in the exometabolite-endometabolite ratios. For both lines, the exudation of succinate was largely unaltered, despite the fact that the levels of this metabolite were increased within the roots of both sets of transgenics. While such shifts were also apparent for some other metabolites in the mMDH lines (e.g. trehalose, Glc, Gal, and Man were exuded at decreased levels, while the relative exudation of Lys was elevated in comparison with the cellular content of the transgenics), the exudation pattern of the FL lines was dramatically divergent from that of the endometabolites of these lines. As described above, the FL lines were characterized by large changes in the levels of both endometabolites and exometabolites. When these profiles are compared, they are highly dissimilar; particularly prominent are the discrepancies in changes in Glu and Gln and the organic acids malate, citrate, and shikimate, all of which have considerable shifts in the ratio of endometabolites to exometabolites, indicating that their exudation is most likely actively promoted in these lines.
Redistribution of Isotope following Incubation of Root Material in [13C]Glc
In order to gain further insight into the root metabolism of the transgenic lines, we next performed feeding experiments in which we incubated root material of both transformants in [13C]Glc for a period between 3 and 5 h and determined the redistribution of heavy label using a modified version of the GC-MS metabolite profiling method described above (Roessner-Tunali et al., 2004; Tieman et al., 2006). Interestingly, the changes in redistribution of isotope were more or less conserved across the lines, with results from this experiment in close agreement with the observed decrease in respiration rate (Table IV). In lines from both sets of transformants, a decreased label redistribution to Glu (significant in all lines with the exception of mMDH8) and fumarate (significant in lines mMDH21 and FL63) was observed. By contrast, some of the changes were transformant specific; for example, the redistribution of isotope to malate was either unchanged or increased in the mMDH lines but was reduced in the FL lines (significantly so in the case of lines FL11 and FL41).
Table IV.
13C enrichment studies in 5-week-old mature root tips of FL and mMDH antisense transgenic lines
Root tips were segmented in 5-mm lengths and incubated for 3 and 5 h in [U-13C]Glc. Values are presented as means ± se of six individual plants per line; values in boldface were determined by the t test to be significantly different (P < 0.05) from the wild type. Negligible enrichment is indicated by “nd.”
| Metabolite | Wild Type | mMDH7 | mMDH8 | mMDH21 | FL11 | FL41 | FL63 |
|---|---|---|---|---|---|---|---|
| μmol fractional carbon enrichment g−1 fresh weight h−1 | |||||||
| Ala (×102) | 3.0 ± 1.0 | 5.0 ± 1.0 | 8.0 ± 3.0 | 3.0 ± 1.0 | 5.0 ± 1.0 | 9.0 ± 2.0 | 4.0 ± 1.0 |
| Asp (×102) | 18.0 ± 8.0 | 4.0 ± 2.0 | 23.0 ± 7.0 | 1.0 ± 1.0 | 20.0 ± 10.0 | 5.0 ± 1.0 | 3.0 ± 1.0 |
| Citrate | 31.7 ± 10.3 | 15.6 ± 4.7 | 69.1 ± 21.5 | 7.3 ± 2.7 | 7.5 ± 2.0 | 5.2 ± 2.6 | 26.1 ± 12.6 |
| Fru | 33.1 ± 15.8 | 53.9 ± 18.3 | 40.7 ± 11.2 | 58.4 ± 20.1 | 50.3 ± 16.8 | 20.3 ± 14.4 | 47.0 ± 15.0 |
| Fumarate | 0.7 ± 0.1 | 0.2 ± 0.2 | 1.1 ± 0.6 | 0.2 ± 0.1 | 0.3 ± 0.2 | 0.5 ± 0.1 | 0.2 ± 0.1 |
| Glu (×102) | 329.0 ± 135.0 | 19.0 ± 8.0 | 95.0 ± 33.0 | 20.0 ± 10.0 | 18.0 ± 8.0 | 7.0 ± 2.0 | 3.0 ± 1.0 |
| Malate | 4.7 ± 0.9 | 3.3 ± 1.8 | 8.9 ± 1.6 | 2.6 ± 0.8 | 0.9 ± 0.3 | 0.5 ± 0.2 | 4.0 ± 1.3 |
| Quinate | 2.0 ± 0.7 | 0.8 ± 0.3 | 1.7 ± 0.5 | 1.4 ± 0.9 | 1.3 ± 0.4 | 0.8 ± 0.3 | 0.9 ± 0.2 |
| Ser | 0.5 ± 0.3 | 0.5 ± 0.2 | 0.8 ± 0.4 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.2 | 0.1 ± 0.0 |
| Val | 0.7 ± 0.4 | 2.3 ± 1.5 | nd | nd | nd | 0.3 ± 0.2 | 0.1 ± 0.0 |
Evaluation of Phytohormone Content and the Expression of Phytohormone-Associated Genes in Roots of the Transgenics
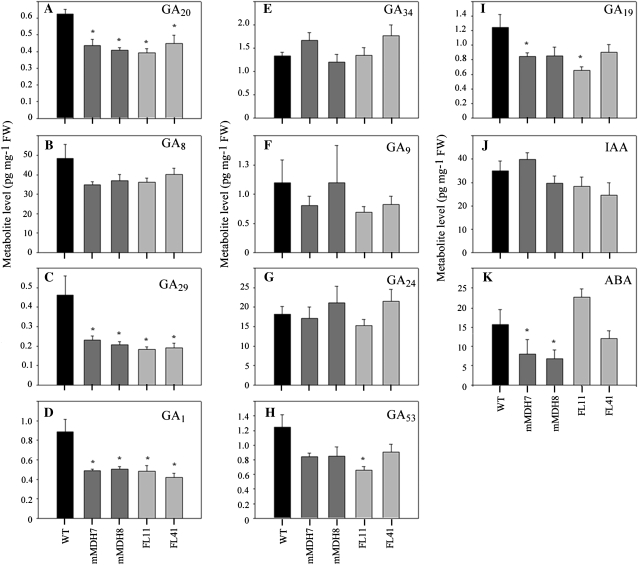
Given the observed alterations in root architecture, we thought it prudent to evaluate the levels of the phytohormones abscisic acid (ABA), auxin (IAA), and gibberellins (GAs), which have all been strongly implicated to have great importance in root development (Yaxley et al., 2001; Wilkinson and Davies, 2002; Swarup et al., 2008; Ubeda-Tomas et al., 2008). For this purpose, we applied two established GC-MS-based protocols to extracts taken from the same samples as used for the metabolite quantification described above. The results revealed that there were relatively few clear trends with respect to the observed morphological phenotypes, with the exception that the general reduction in the level of C19 GAs (GA20, GA29, and GA1 decreased in all transgenic lines tested, while there were additionally significant decreases in the C20-GAs GA53 and GA19 in a subset of the lines), is consistent with the general reduction of root biomass in both sets of transgenics (Fig. 3). By contrast, the level of auxin was invariant in the transgenics, while that of ABA was decreased in the mMDH lines but unaltered in the FL lines (Fig. 3).
Figure 3.
Phytohormone profiling of mMDH 7, mMDH8, FL11, and FL41 lines with 5-week-old plants. Quantifications include the measurement of GA (A–I), IAA (J), and ABA (K). The values are means ± se of six individual plants. Asterisks indicate values determined by the t test to be significantly different from the wild-type (WT) control (P < 0.05). FW, Fresh weight.
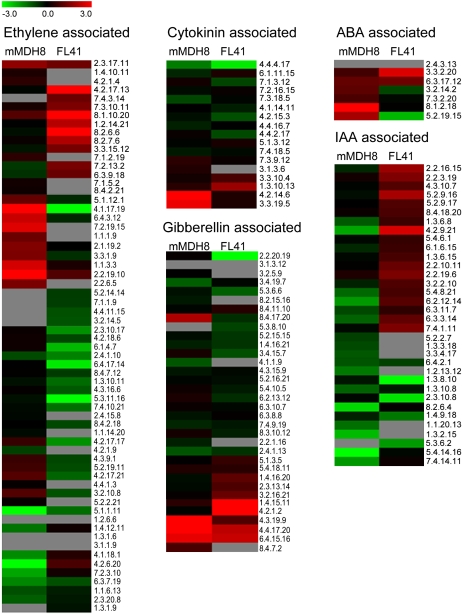
Despite the relative paucity of changes in the levels of the hormones themselves, we next decided to evaluate the expression of genes associated with hormone perception or signal transduction relay. In order to achieve this, we isolated RNA from the same material used for the phytohormone measurements, with the exception that only one representative transgenic line was considered, and evaluated gene expression using the TOM1 microarray (Alba et al., 2004). The pertinent data from this transcript profiling study is presented in Figure 4, with transcripts being grouped on the basis of their MapMan ontologies and detailed description of their identities being provided in Supplemental Table S4. While this presentation revealed a large number of changes, these generally did not correspond to the observed similarities or differences in phenotype across the lines. Given that the phenotypes displayed by both mMDH and fumarase were relatively dramatic and were either conserved or opposite, we had postulated that we may see a similar behavior at the transcript level of key genes. Studying Figure 4, however, revealed only a few putative homologous genes displaying antagonistic expression in the transgenics, for example, auxin response factor 3 (EC 4.2.9.21), ABA-responsive protein (EC 5.2.19.15), and MADS box transcription, and a few that displayed similar changes in gene expression, for example, GA 3-oxidase 1 (EC 6.4.15.16) GA 2-oxidase (EC 4.4.17.20), and the response regulator protein (EC 4.4.4.17).
Figure 4.
Transcript responses involved in regulatory and/or signaling responses in root growth and development. The values are representative of the log2-transformed values of three individual hybridizations, statistically analyzed and Loewess tip-point normalized in R, and classed according to MapMan classifications (Thimm et al., 2004). Numerical annotations refer to the array layout, and putative identifications are specified in Supplemental Table S4.
Analysis of the Influence of Modified Shoot Metabolism on Root Function in the Transgenics
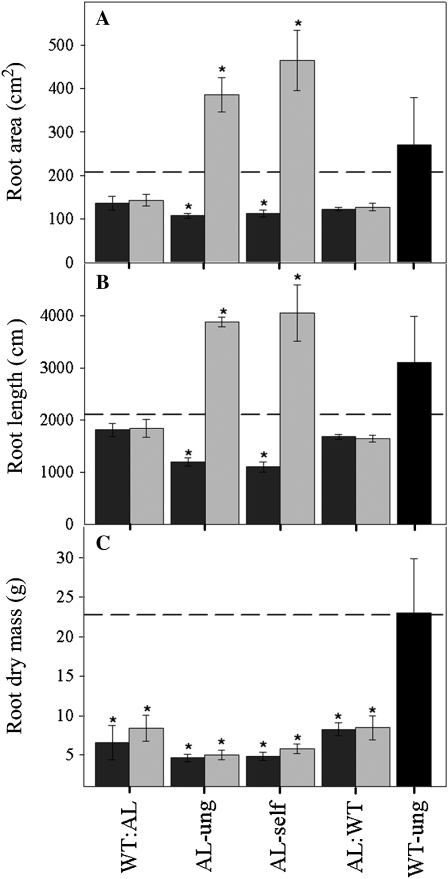
Given that the majority of the experiments described thus far have focused directly on the roots (or root system), we decided to additionally evaluate basic aspects of root morphology and metabolism from the whole plant perspective. For this purpose, reciprocal grafting experiments between transgenic lines (mMDH8 and FL41) and wild-type controls were performed and root morphological parameters of the grafted material were determined. Plants were grown on vermiculite for a period of 4 weeks, after which graftings were carried out between wild-type stock and transgenic scions and vice versa. The resultant grafts were then left for another 2 weeks prior to the determination of morphological attributes. The grafting of wild-type scions onto transgenic stock was able to revert the transgenic root-length and root-area phenotypes to resemble those of the wild type (i.e. they resulted in a relative reduction in the length of roots in the FL line and a relative increase in the mMDH line; Fig. 5, A and B). By contrast, the grafts were not able to alter the root biomass phenotype of either line, indicating that this parameter was independent of changes in the phenotype of the source organs (Fig. 5C). For all morphological parameters under consideration, wild type-to-wild type grafts were not significantly different from the untreated wild type, allowing us to exclude that the effects we observed were a direct consequence of the grafting process per se.
Figure 5.
Root morphology of 6-week-old grafted scions. Grafting the wild-type leafstock onto transgenic rootstock (either mMDH8 [dark gray bars] or FL41 [light gray bars]) or the reciprocal grafting led to a restoration in both root area (A) and total root length (B) to those of the wild-type self-grafted (WT:WT) control (dashed horizontal line). These parameters were significantly altered (reduced in mMDH8 and increased in FL41) in the ungrafted (ung) and self-grafted (self) antisense lines (AL). However, the grafting could not restore the root dry mass (C). Values are means ± se of five individual grafting combinations. Asterisks indicate values determined by the t test (P < 0.05).
In a second experiment, we repeated the grafting but only with the most extreme transgenic line, and this time we harvested root tips from the rootstock of the grafted plants and determined their metabolite content by GC-MS (Table V). As would perhaps be expected, the metabolite levels of the untreated transgenic lines were highly consistent with those reported in Supplemental Table S2, while the reciprocal grafts contained far fewer metabolic changes than observed in the ungrafted transgenic plants (irrespective of the direction of the graft). Evaluating the entire metabolite data sets by hierarchical cluster analysis (Saeed et al., 2003) revealed that the ungrafted FL metabolome is the most divergent from that of the ungrafted wild type. However, close inspection of the data in Table V reveals specific patterns of metabolic change that can be discerned that may contribute to the phenotypic restoration. Using this approach, we were able to identify that the levels of β-Ala and Pro were significantly different in the ungrafted roots of both transgenic lines but returned to wild-type levels following grafting of a wild-type scion. Using this approach, other subsets of metabolites could be identified that responded to the morphological restoration associated with grafting in one or other of the transgenic lines. Analysis of these data clearly revealed that levels of the metabolites glycerol and glycerol-3 phosphate were altered by the grafting process itself.
Table V.
Metabolite profiles of grafting combinations of transgenic scion with wild-type rootstock (MDH:WT, FL:WT), wild-type scion with transgenic rootstock (WT:MDH, WT:FL), wild-type self-grafting (WT:WT), or ungrafted entities (wild type, MDH, FL)
Data are normalized with respect to the mean response calculated for the wild type. Values presented are means ± se of four replicates; values in boldface were determined by the t test to be significantly different (P < 0.05). nd, Not detected.
| Metabolite | Wild Type | mMDH8 | FL41 | MDH:WT | FL:WT | WT:MDH | WT:FL | WT:WT |
|---|---|---|---|---|---|---|---|---|
| Ala | 1.00 ± 0.32 | 1.08 ± 0.27 | 1.81 ± 0.24 | 0.81 ± 0.33 | 0.38 ± 0.13 | 0.85 ± 0.10 | 2.29 ± 0.89 | 0.88 ± 0.26 |
| β-Ala | 1.00 ± 0.09 | 1.89 ± 0.36 | 3.30 ± 0.17 | 2.62 ± 1.15 | 0.62 ± 0.10 | 0.85 ± 0.09 | 1.12 ± 0.16 | 1.05 ± 0.23 |
| Asn | 1.00 ± 0.55 | 1.44 ± 0.33 | 2.64 ± 0.23 | 0.46 ± 0.21 | 0.94 ± 0.46 | 0.97 ± 0.66 | 2.80 ± 0.60 | 0.33 ± 0.11 |
| Asp | 1.00 ± 0.17 | 1.39 ± 0.30 | 2.96 ± 0.18 | 0.52 ± 0.22 | 1.04 ± 0.15 | 0.90 ± 0.19 | 1.18 ± 0.01 | 0.82 ± 0.16 |
| Benzoate | 1.00 ± 0.14 | 0.79 ± 0.17 | 1.77 ± 0.14 | 0.62 ± 0.10 | 0.82 ± 0.11 | 0.86 ± 0.05 | 1.04 ± 0.12 | 0.97 ± 0.13 |
| GABA | 1.00 ± 0.29 | 0.76 ± 0.34 | 1.76 ± 0.35 | 0.97 ± 0.29 | 1.20 ± 0.19 | 1.39 ± 0.21 | 1.61 ± 0.12 | 1.10 ± 0.24 |
| Citrate | 1.00 ± 0.19 | 1.06 ± 0.31 | 2.76 ± 0.21 | 0.13 ± 0.02 | 1.07 ± 0.07 | 0.73 ± 0.19 | 0.74 ± 0.25 | 0.72 ± 0.22 |
| Fru | 1.00 ± 0.02 | 0.87 ± 0.30 | 1.30 ± 0.23 | 0.07 ± 0.03 | 0.88 ± 0.07 | 0.82 ± 0.17 | 0.90 ± 0.03 | 1.02 ± 0.02 |
| Fru 6-P | 1.00 ± 0.31 | 1.02 ± 0.42 | 2.49 ± 0.45 | 2.12 ± 0.95 | 1.04 ± 0.12 | 0.74 ± 0.19 | 1.55 ± 0.21 | 0.75 ± 0.12 |
| Fumarate | 1.00 ± 0.09 | 1.21 ± 0.22 | 2.31 ± 0.19 | 2.12 ± 0.94 | 0.58 ± 0.11 | 0.49 ± 0.10 | 0.82 ± 0.11 | 1.04 ± 0.38 |
| Galacturonate | 1.00 ± 0.26 | 0.97 ± 0.51 | 3.05 ± 0.52 | 0.68 ± 0.30 | 1.39 ± 0.06 | 1.22 ± 0.16 | 1.89 ± 0.22 | 1.14 ± 0.19 |
| Glc | 1.00 ± 0.65 | 0.79 ± 0.27 | 2.35 ± 0.26 | 0.08 ± 0.04 | 3.95 ± 0.83 | 0.58 ± 0.54 | 0.79 ± 0.53 | 1.56 ± 0.56 |
| Glc 6-P | 1.00 ± 0.29 | 1.07 ± 0.41 | 2.39 ± 0.42 | 2.05 ± 0.91 | 1.03 ± 0.10 | 0.76 ± 0.20 | 1.66 ± 0.22 | 0.73 ± 0.14 |
| Gln | 1.00 ± 0.24 | 0.90 ± 0.35 | 4.31 ± 0.24 | 0.09 ± 0.04 | 1.06 ± 0.23 | 1.07 ± 0.25 | 1.44 ± 0.01 | 0.85 ± 0.25 |
| Glycerate | 1.00 ± 0.15 | 1.26 ± 0.23 | 2.27 ± 0.16 | 2.81 ± 1.22 | 1.18 ± 0.14 | 1.32 ± 0.20 | 2.09 ± 0.24 | 1.07 ± 0.24 |
| Glycerol | 1.00 ± 0.20 | 1.11 ± 0.19 | 2.80 ± 0.11 | 2.15 ± 0.78 | 1.25 ± 0.30 | 2.19 ± 0.53 | 2.72 ± 0.72 | 2.47 ± 0.48 |
| Glycerol 3-P | 1.00 ± 0.27 | 1.30 ± 0.32 | 2.31 ± 0.25 | 1.00 ± 0.44 | 1.38 ± 0.23 | 2.22 ± 0.78 | 4.05 ± 1.19 | 2.73 ± 0.91 |
| Gly | 1.00 ± 0.12 | 1.62 ± 0.30 | 3.17 ± 0.12 | 0.76 ± 0.31 | 1.17 ± 0.17 | 1.12 ± 0.12 | 1.67 ± 0.41 | 1.52 ± 0.54 |
| His | 1.00 ± 0.27 | nd | nd | 0.26 ± 0.12 | 0.89 ± 0.20 | 0.90 ±.0.45 | 2.17 ± 0.32 | 1.98 ± 1.04 |
| Myoinositol | 1.00 ± 0.10 | 0.64 ± 0.48 | 1.83 ± 0.38 | 0.56 ± 0.21 | 1.13 ± 0.03 | 1.14 ± 0.02 | 1.09 ± 0.01 | 0.82 ± 0.12 |
| Ile | 1.00 ± 0.16 | 1.19 ± 0.36 | 2.04 ± 0.20 | 0.90 ± 0.38 | 1.67 ± 0.24 | 1.91 ± 0.83 | 2.33 ± 0.58 | 1.34 ± 0.36 |
| Lys | 1.00 ± 0.18 | 0.78 ± 0.51 | 1.66 ± 0.44 | 0.64 ± 0.27 | 1.14 ± 0.19 | 1.01 ± 0.27 | 1.69 ± 0.18 | 1.01 ± 0.21 |
| Malate | 1.00 ± 0.19 | 1.37 ± 0.31 | 2.55 ± 0.32 | 0.57 ± 0.20 | 1.05 ± 0.08 | 0.86 ± 0.21 | 1.19 ± 0.01 | 0.93 ± 0.11 |
| Maltitol | 1.00 ± 0.25 | 1.38 ± 0.25 | 3.37 ± 0.37 | 1.75 ± 0.77 | 1.94 ± 0.26 | 1.29 ± 0.24 | 2.31 ± 0.25 | 1.38 ± 0.44 |
| Maltose | 1.00 ± 0.35 | 1.36 ± 0.36 | 2.04 ± 0.27 | 2.17 ± 0.90 | 1.41 ± 0.20 | 1.08 ± 0.18 | 1.94 ± 0.30 | 1.06 ± 0.14 |
| Man | 1.00 ± 0.09 | 1.00 ± 0.19 | 2.63 ± 0.30 | 0.03 ± 0.01 | 1.15 ± 0.13 | 0.92 ± 0.22 | 1.79 ± 0.35 | 1.10 ± 0.17 |
| Met | 1.00 ± 0.25 | 1.03 ± 0.27 | 1.99 ± 0.27 | 0.55 ± 0.23 | 1.02 ± 0.24 | 0.86 ± 0.25 | 1.90 ± 0.26 | 1.11 ± 0.32 |
| Phe | 1.00 ± 0.21 | 1.03 ± 0.26 | 1.93 ± 0.24 | 0.73 ± 0.31 | 1.15 ± 0.14 | 1.05 ± 0.23 | 1.65 ± 0.20 | 1.19 ± 0.31 |
| Pro | 1.00 ± 0.15 | 2.15 ± 0.34 | 2.23 ± 0.34 | 1.99 ± 0.99 | 1.05 ± 0.15 | 0.93 ± 0.25 | 1.31 ± 0.24 | 0.76 ± 0.13 |
| Putrescine | 1.00 ± 0.12 | 0.67 ± 0.38 | 2.44 ± 0.46 | 0.17 ± 0.07 | 0.36 ± 0.07 | 0.69 ± 0.12 | 0.51 ± 0.06 | 0.92 ± 0.24 |
| Raffinose | 1.00 ± 0.38 | 3.62 ± 0.39 | 1.95 ± 0.23 | 0.18 ± 0.08 | 0.27 ± 0.02 | 0.25 ± 0.03 | 0.34 ± 0.05 | 0.25 ± 0.05 |
| Saccharate | 1.00 ± 0.20 | 0.83 ± 0.23 | 2.15 ± 0.21 | 0.62 ± 0.25 | 1.20 ± 0.03 | 1.23 ± 0.04 | 1.04 ± 0.04 | 1.12 ± 0.08 |
| Ser | 1.00 ± 0.43 | 2.25 ± 0.42 | 2.76 ± 0.24 | 1.20 ± 0.50 | 0.63 ± 0.08 | 0.58 ± 0.28 | 1.23 ± 0.29 | 0.60 ± 0.14 |
| Succinate | 1.00 ± 0.17 | 2.42 ± 0.29 | 1.20 ± 0.23 | 2.93 ± 1.27 | 0.88 ± 0.08 | 1.21 ± 0.11 | 1.08 ± 0.15 | 0.99 ± 0.17 |
| Suc | 1.00 ± 0.33 | 2.30 ± 0.56 | 1.42 ± 0.41 | nd | 0.80 ± 0.21 | 0.71 ± 0.15 | 1.50 ± 0.17 | 0.84 ± 0.28 |
| Thr | 1.00 ± 0.49 | 1.82 ± 0.37 | 2.36 ± 0.23 | 0.75 ± 0.31 | 0.78 ± 0.11 | 0.54 ± 0.17 | 0.85 ± 0.11 | 0.58 ± 0.09 |
| Threonate | 1.00 ± 0.49 | 0.72 ± 0.20 | 1.92 ± 0.26 | 0.75 ± 0.31 | 0.78 ± 0.11 | 0.54 ± 0.17 | 0.85 ± 0.11 | 0.58 ± 0.09 |
| Tyr | 1.00 ± 0.26 | 0.71 ± 0.46 | 1.52 ± 0.43 | 0.17 ± 0.07 | 1.00 ± 0.29 | 1.13 ± 0.21 | 1.38 ± 0.39 | 1.94 ± 0.48 |
| Trehalose | 1.00 ± 0.13 | 0.51 ± 0.42 | 1.59 ± 0.19 | 0.29 ± 0.13 | 0.68 ± 0.04 | 0.81 ± 0.16 | 0.99 ± 0.23 | 1.12 ± 0.28 |
| Trp | 1.00 ± 0.17 | 0.90 ± 0.49 | 1.34 ± 0.53 | 0.01 ± 0.00 | 1.07 ± 0.29 | 0.78 ± 0.12 | 0.60 ± 0.05 | 0.64 ± 0.15 |
| Val | 1.00 ± 0.10 | 1.12 ± 0.30 | 1.90 ± 0.18 | 0.91 ± 0.38 | 1.04 ± 0.17 | 0.95 ± 0.20 | 1.43 ± 0.17 | 1.21 ± 0.26 |
| Xylitol | 1.00 ± 0.22 | 1.06 ± 0.31 | 2.41 ± 0.26 | 0.16 ± 0.05 | 0.88 ± 0.11 | 1.13 ± 0.20 | 1.83 ± 0.46 | 1.74 ± 0.54 |
DISCUSSION
The aim of this work was to determine the importance of enzymes of malate metabolism for root morphology and function. Initial observations previously suggested that root biomass was decreased in transgenic tomato, exhibiting decreased expression either of the mitochondrial malate dehydrogenase or the predominantly expressed isoform of fumarase. On evaluation of root metabolism, in plants prior to flowering, we confirmed the reduction in root biomass in both sets of lines. In addition, we observed that the mMDH lines displayed decreased root length and a decreased exudate pH (whereas the FL lines displayed slightly increased root length). We also documented that, as would be expected, the rate of respiration in the roots was more severely compromised than in the leaves of these lines. This finding is interesting as it suggests that there is less flexibility in respiration in heterotrophic organs; however, currently we can only speculate about the reasons underlying it. It is highly likely that this observation merely reflects the relative importance of the TCA cycle in the various organs; however, we cannot formally exclude that alternative respiratory substrates are more efficiently used in the leaf. In keeping with the first hypothesis are the facts that the respiration rates documented here in the roots of the wild type (Fig. 1I) are far in excess of those reported previously for wild-type tomato leaves (Nunes-Nesi et al., 2005, 2007a) and that the rate of the TCA cycle in the illuminated leaf is, at least to some extent, restricted (Tcherkez et al., 2005; Nunes-Nesi et al., 2007b). In addition, the results of the 13C-labeling experiments (Table IV) reinforce this, since the results from tomato root are suggestive of higher fluxes than those previously reported for tomato leaves (Studart-Guimarães et al., 2007). Furthermore, a severe inhibition of respiration is also evidenced by the reduced levels of adenylates observed in the transgenic lines (Fig. 2). Moreover, there is no current evidence to suggest that TCA cycle bypasses do not function as effectively in roots as in leaves (Ishizaki et al., 2005; Nunes-Nesi et al., 2005). Indeed, the facts that the levels of reductant are not dramatically reduced and that the level of ascorbate is even enhanced in the transgenic lines (Fig. 2) are highly suggestive that these bypasses are indeed engaged in the transformants. That said, irrespective of the exact reason behind the observed differences, it is clear that the TCA cycle plays a less redundant role in root metabolism than in leaf metabolism.
A major goal of this work was to improve our understanding of the influence of malate metabolism on root function in the tomato. Particular attention was paid to dissecting whether the restriction of root biomass observed here was the consequence of altered source function, altered root exudation, or merely an effect of altering metabolism in the root. In order to address this question, we took a range of approaches, including metabolite profiling of both cellular (endo-) and exuded (exo-) metabolites of the root system as well as analyzing the levels of phytohormones and the expression of genes associated with their perception and/or signal relay. For this purpose, we adopted an established GC-MS protocol to evaluate endometabolite and exometabolite contents of the transgenic root systems as well as the endometabolite contents in the root tips of grafted plants and used targeted approaches to measure phytohormones, carbohydrates, adenylates, and redox compounds in the root tip. GC-MS analysis of the endometabolites revealed very few changes in the mMDH lines, while the majority of endometabolites decreased in the FL lines. Comparison of the changes in exometabolite contents in the FL and mMDH lines at the end of the light period revealed that exudation was also markedly different between the lines. The levels of a wide range of exometabolites were elevated in the FL lines, while there was little change in the mMDH lines. These results provide compelling evidence that the reduced root biomass phenotype is not a consequence of a reduced organic acid exudation in the transgenics. This observation is somewhat at odds with previous studies showing that manipulations of TCA cycle metabolism in the roots can greatly improve fitness via a root exudate-mediated mechanism (Koyama et al., 2000; López-Bucio et al., 2000, 2003). Several factors could explain this discrepancy, including the facts that we are studying different species, subject to different nutrition and genetically modified at a different reaction step. However, irrespective of the reason underlying the different conclusions of these studies, our work clearly suggests that the change in biomass observed is not the consequence of a reduced exudation of organic acids.
The fact that one of the sets of transgenics was previously demonstrated to display an enhanced rate of photosynthesis and the other displayed a decreased rate (Nunes-Nesi et al., 2005, 2007a) suggests that the root biomass is unlikely to be greatly influenced by the aerial part of the plant. The results of reciprocal grafting experiments confirmed our intuition here, since although a wild-type scion could restore the transgenic stock root length and area to wild-type values, it had little effect on the root biomass. The recovery of root length and root area following grafting of the wild-type scion to the transgenic stock is highly interesting and suggests that these parameters are dramatically influenced by photoassimilate availability. However, the total root biomass and root architecture appear to be regulated largely independently of this. Results of a recent elegant study reveal that root tip contact with low-phosphate medium reprograms root architecture (Svistoonoff et al., 2007). Similar effects have also been documented for growth on low-nitrate medium (Zhang and Forde, 1998; Forde and Lea, 2007). The fact that the transgenic lines studied here exhibit altered architecture implies that the altered exudation profiles may play a role in nutrient uptake; however, analysis of a broader range of lines than were examined in this study would be required in order to confirm this hypothesis.
What the results from these studies do clearly demonstrate, however, is that the reduced root biomass is due to a root-specific effect. One possible cause for this is the dramatic reduction in respiration in this organ. This reduction can be seen in both the direct oxygen consumption measurements and the results obtained from the 13C isotope-labeling experiments, which imply a slower general metabolism in the transformants. This trend, however, is not apparent in the steady-state levels of endometabolites, presumably due to the different exudation rates of the different transgenics. That the rate of respiration and the reduced root biomass correlate, taken alongside the previously observed links between energy metabolism, biosynthesis, and growth (Regierer et al., 2002; Carrari et al., 2003; Devaux et al., 2003), strongly implies that the reduction in root biomass is the consequence of an impairment in the root's own energy metabolism. While decreased provision of the necessary precursors and energy for biosynthesis in the transgenics seems the logical mechanism by which to explain the growth inhibition of the roots, we also need to take into account the results of the phytohormone profiling. While there were differences in the expression levels of a relatively large number of genes associated with phytohormones and their downstream regulatory cascades, it is important to note that the only phytohormones that displayed consistent changes across the transgenic lines were GAs. Perhaps equally importantly, we identified no antagonistic changes in the levels of phytohormones themselves and hardly any antagonistic changes in the levels of transcripts associated with their signal perception/relay, suggesting that they are unlikely to be involved in the establishment of the defined altered root morphological phenotypes displayed by the transgenic lines. This finding is somewhat surprising given the wealth of literature indicating an important role of phytohormones in root development. Several studies have either directly or indirectly suggested important roles of auxin and/or ethylene in regulating root size in the elongation zone (Chilley et al., 2006; Fitzgerald et al., 2006; Rahman et al., 2007; Stepanova et al., 2007; Okamoto et al., 2008), while others pinpoint an important role of cytokinin in the control of root cell division (Riefler et al., 2006). These findings are largely in congruence with previous observations of the roles of phytohormones in the control of root growth and architecture (Malamy, 2005; for review, see López-Bucio et al., 2006).
From our current study, we can state that the steady-state levels of these compounds in the root per se do not influence root branching and other morphological effects observed in the transgenics. We cannot, however, formally exclude that control of root development could be mediated by the influx of phytohormones from aerial parts of the plant, since several of these phenotypes were rescued by the grafting of a wild-type scion onto the transgenic rootstock. While we were able to demonstrate that this was able, at least partially, to complement the metabolic phenotype of the transgenics, we did not determine if this resulted in an altered exchange rate of phytohormones between the aerial and subterranean parts of the plant. Our results, however, do allow us to conclude that, despite the presence of compelling evidence for roles for IAA and ABA in mediating root development (Grieneisen et al., 2007; de Grauwe et al., 2008; Swarup et al., 2008, they are involved in determining the root biomass phenotype described here. The same, however, cannot be said of the changes observed concerning GA. Recent studies in pea (Pisum sativum) mutants deficient in gibberellin revealed that their dwarf phenotype could be complemented via provision of the hormone (Yaxley et al., 2001). Reverse genetic analysis of tobacco (Nicotiana tabacum) GA 2-oxidase, however, which was strongly down-regulated in our transgenics, revealed that deficiency of this enzyme resulted in shunted aerial growth and a much reduced lignin deposition (Biemelt et al., 2004). Given that such phenotypic characterizations have also been imposed by the modification of GA metabolism, we cannot rule out the possibility that the mechanism by which restriction of the TCA cycle results in modified root biomass is at least in part mediated by changes in the hormones that are apparent in these lines. While the precise nature of the interaction between energy metabolism and GA-mediated control of growth could not be resolved in this study, it remains an exciting topic for future research.
MATERIALS AND METHODS
Plant Material
Previously characterized tomato (Solanum lycopersicum ‘Moneymaker’) seeds, exhibiting reduced expression of mitochondrial malate dehydrogenase and fumarase (Nunes-Nesi et al., 2005, 2007a), were sterilized and germinated on 2× MS medium (Murashige and Skoog, 1962). Plants were grown in a long-day regime at day/night temperatures of 22°C/20°C and a relative humidity of 50% on a vermiculite medium supplemented with slow-release fertilizer (Lewatit HD 50; Beyer). The presence of the transgene was verified by PCR using standard protocols prior to analysis of enzyme activities of the protein in question. Rootstock-to-leafstock wedge grafts were performed on 4-week-old tomato plants that were defoliated to the third fully expanded leaf, with V-intersection grafts performed and pinned by sterile aluminum supports. Plants were covered with 20-L clear plastic bags and allowed to adjust to natural lighting conditions for 24 h, after which they were returned to the growth conditions described above.
Chemicals
Unless stated otherwise, all chemicals, cofactors, and enzymes were purchased from either Sigma-Aldrich or Merck.
Exudation
Root exometabolites were collected at the end of the light period (unless stated otherwise) by gently excavating the root system from the vermiculite growth medium, washing, and incubating in double distilled water (pH 6.8) under gentle agitation (100 rpm) for 1 h.
Bacterial Culturing Methods
The presence of colony-forming units was determined in order to monitor microbial activity exactly as defined previously (Weisskopf et al., 2005), and a value of loge < 0.69 was deemed satisfactory.
Enzyme Analyses
Enzymes were extracted as described previously (Tauberger et al., 2000) and assayed exactly as described in the literature (Gibon et al., 2004; Nunes-Nesi et al., 2005).
Immunodetection of Fumarase Protein
Protein-blot analysis of fumarase protein was carried out on 33 μg of crude protein extract according to Nunes-Nesi et al. (2007a) and subjected to protein-blot analysis according to standard procedures. Membranes were probed with primary rabbit antibody raised against Arabidopsis (Arabidopsis thaliana) fumarase protein (Behal and Oliver, 1997). The secondary antibody was an affinity-purified IR Dye800-conjugated goat anti-rabbit antibody (Rockland Immunochemicals). Signals were quantified using an Odyssey Infrared Imager system (Li-Cor Biosciences).
Metabolic Profiling
Cellular metabolite levels were analyzed as outlined by Roessner et al. (2001), with the exceptions that initial parameters taken were optimized for tomato following the method of Roessner-Tunali et al. (2003) and that root tissue was homogenized with a mortar and pestle in liquid nitrogen. Identification and quantification were optimized for tomato root tissue with known standards, and the spectrum was adjusted accordingly. In the case of root exometabolites, these were collected in double distilled water (pH 6.8) by gentle agitation of roots (for 1 h) following excavation from the vermiculite medium. Unless otherwise indicated, excavation was carried out at the end of the light period. All material collected was rapidly frozen in liquid nitrogen and stored at −80°C until further analysis. Optimization procedures for determination of the exudate metabolite levels were carried out as described above for the cellular metabolites of tomato root samples.
Carbohydrate, Energy, and Redox Metabolite Measurements
The levels of starch, Suc, Fru, and Glc in the leaf tissue were determined exactly as described previously (Müller-Röber et al., 1992).The procedure of extraction and assay of NADs was performed according to the method described by Gibon and Larher (1997). Extraction and spectrophotometric determinations for NAD(P), NAD(P)H, ascorbate, and glutathione were performed exactly as described by Queval and Noctor (2007).
[13C]Glc Feeding
Isotopic feeding of root material was done following the protocol outlined by Roessner-Tunali et al. (2004). In brief, mature roots were cut into 5-mm segments and washed three times in cold 10 mm MES-KOH (pH 6.5), and approximately 0.2 g of tissue was incubated either in 20 mm [U-13C]Glc (Omicron Biochemicals) or 20 mm natural abundance Glc in a total volume of 5 mL of 10 mm MES-KOH buffer (pH 6.5) per 100-mL Erlenmeyer flask. The flasks were incubated for 3 and 5 h, respectively, under an agitation of 100 rpm. After this time period, tissue was thoroughly washed with 10 mm MES-KOH buffer (pH 6.5), rapidly blotted dry on tissue paper, and snap frozen in liquid nitrogen. Frozen tissue was stored at −80°C until further analysis. Labeling patterns were analyzed as described previously (Giegé et al., 2003; Roessner-Tunali et al., 2004).
Transcript Profiling
Total RNA extraction for 5-week-old root material was done using Trizol according to the manufacturer's specifications. Following this, first-strand cDNA synthesis was carried out using the SuperScript indirect cDNA labeling kit (Invitrogen). Typically, 15 to 20 μg of total RNA (DNase I treated) was incubated at 70°C for 10 min with 5 μg of oligo(dT)20 primer in a total volume of 18 μL and placed on ice for 5 min. Subsequently, a master mix consisting of 5× first-strand buffer, 0.15 μmol of dithiothreitol, 15 mmol of deoxynucleoside triphosphate mix, RNAseOUT, and SuperScript III reverse transcriptase was added and the tube was incubated at 46°C. The mixture was hydrolyzed with 1 n NaOH, thoroughly mixed, and incubated at 70°C for 10 min, after which it was neutralized with 1 n HCl. Sodium acetate (3 m, pH 5.2) was added before proceeding to first-strand cDNA purification, following the kit instructions. Labeling and hybridization of the TOM1 array were followed directly as described previously (Alba et al., 2004). The TOM1 arrays (Cornell University) were hybridized and scanned with a Fuji MAS FLA-8000 microarray scanner. The GeneSpotter software (MicroDiscovery) was used for the grid positioning and signal quantification. The resulting data were analyzed using the LIMMA package (Smyth and Speed, 2003) for the bioconductor software (Gentleman et al., 2004). Data were normalized using within-array print-tip Loess and between-array quantile normalization. P values were corrected using a false discovery rate correction (Benjamini and Hochberg, 1995), and a false discovery rate P value (q value) of <0.05 was deemed significant. Data were visualized with PageMan software (Usadel et al., 2006).
Root Respiration
Root tips (25 mg) were cut with a sharp blade, washed, and incubated in 10 mm MES-KOH (pH 6.5) in a Clark-type oxygen electrode. The rate of oxygen consumption was calculated as described by Geigenberger et al. (2000).
Root Phenotype Assessment
Root phenotypes were digitally obtained and analyzed with Rootedge software (version 2.3; Iowa State University Research Foundation) as described by Kaspar and Ewing (1997). Cell size measurements were performed on root sections.
Phytohormone Determination
Determination of abscisic acid was performed essentially as described by Peng et al. (1999). In brief, frozen samples (0.5 g) were homogenized and extracted overnight in 20 mL of 80% methanol. After extraction, each sample was reduced in vacuo and diluted with 20 mL of water. The aqueous phase was adjusted to pH 2.8 with 1 m HCl and partitioned four times with equal volumes of ethyl acetate. The ethyl acetate extracts were combined and evaporated to dryness. The residue was dissolved in 1 mL of 10% methanol and applied to a preequilibrated C18 cartridge (Waters; http://www.waters.com). The column was washed with aqueous acetic acid (pH 3.0), and then ABA was eluted with 80% methanol. After evaporation to dryness, the samples were derivatized as described (Lisec et al., 2006) and analyzed by comparison with authentic standards using GC-MS. IAA and GAs were analyzed by GC-MS exactly as described previously (Björklund et al., 2007).
Statistical Analyses
Unless otherwise specified, statistical analyses were performed using the t test embedded in the Microsoft Excel software. Only the return of P < 0.05 was designated significant.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Relative response ratios of diurnal-like and extended dark period of root metabolites from 5-week-old wild-type tomato roots.
Supplemental Figure S2. Relative response ratios of diurnal-like and extended dark period of exudates collected from 5-week-old wild-type tomato roots.
Supplemental Table S1. Relative root exudates of antisense FL and mMDH lines.
Supplemental Table S2. Absolute endometabolite concentrations (mmol g−1 fresh weight) of antisense FL and mMDH lines.
Supplemental Table S3. Absolute exudate concentrations (mmol g−1 fresh weight) of antisense FL and mMDH lines.
Supplemental Table S4. Putative identifications of transcripts involved in phytohormone responses, as identified from the corresponding MapMan annotation files (Thimm et al., 2004), and numerically listed in Figure 4.
Supplementary Material
Acknowledgments
We thank Drs. Matthew Hannah and Anna Lytovchenko (Max Planck Institute of Molecular Plant Physiology) for advice on grafting and proofreading, respectively. We thank Ingabritt Carlsson for help with IAA and GA analysis. We are also indebted to Dr. Karin Köhl and Helga Kulka (Max Planck Institute of Molecular Plant Physiology) for taking excellent care of the plants.
This work was supported by the Max-Planck-Gesellschaft (M.J.v.d.M., A.N.-N., A.R.F.), the South African National Research Foundation and the Deutscher Akademischer Austausch Dienst (M.J.v.d.M.), the Deutsch Forschungsgemeinschaft (S.O.), and the Swedish Research Council (T.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alisdair R. Fernie (fernie@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D'Ascenzo M, Gordon JS, Rose JK, et al (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39 697–714 [DOI] [PubMed] [Google Scholar]
- Anekonda TS (2001) Extreme growth phenotypes of trees are caused by differences in energy metabolism. Thermochim Acta 373 125–132 [Google Scholar]
- Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ (2003) Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 132 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers H (1961) Respiratory Metabolism in Plants. Row Peterson, Evanston, IL
- Behal RH, Oliver DJ (1997) Biochemical and molecular characterization of fumarase from plants: purification and characterization of the enzyme—cloning, sequencing and expression of the gene. Arch Biochem Biophys 165 397–406 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57 289–300 [Google Scholar]
- Bertin C, Yang X, Weston LA (2003) The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256 67–83 [Google Scholar]
- Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B (2007) Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J 52 499–511 [DOI] [PubMed] [Google Scholar]
- Budde RJA, Randall DD (1990) Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light-dependent manner. Proc Natl Acad Sci USA 87 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger Y, Sa'ar U, Distelfeld A, Katzir N, Yeselson Y, Shen S, Schaffer AA (2003) Development of sweet melon (Cucumis melo) genotypes combining high sucrose and organic acid content. J Am Soc Hortic Sci 128 537–540 [Google Scholar]
- Carrari F, Coll-Garcia D, Schauer N, Lytovchenko A, Palacios-Rojas N, Balbo I, Rosso M, Fernie AR (2005) Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol 137 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilley PM, Casson SA, Tarkowski P, Hawkins N, Wang KLC, Hussey PJ, Beale M, Ecker JR, Sandberg GK, Lindsey K (2006) The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signalling. Plant Cell 18 3058–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer MD, Titus CHA (2001) Elevated root zone dissolved inorganic carbon can ameliorate aluminium toxicity in tomato seedlings. New Phytol 152 29–39 [DOI] [PubMed] [Google Scholar]
- de Grauwe L, Chaerle L, Dugardeyn J, Decat J, Rieu I, Vriezen WH, Moritz T, Beemster GTS, Phillips AL, Harberd NP, et al (2008) Reduced gibberellin response affects ethylene biosynthesis and responsiveness in the Arabidopsis gai eto2-1 double mutant. New Phytol 177 128–141 [DOI] [PubMed] [Google Scholar]
- de la Fuente JM, Ramírez-Rodríguez V, Cabrera-Ponce JL, Herrera-Estrella L (1997) Aluminium tolerance in transgenic plants by alteration of citrate synthesis. Science 276 1566–1568 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101 15249–15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses GG, Kopka J, Udvardi MK (2005) Lotus japonicus metabolic profiling: development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol 137 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB (2006) Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18 2314–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C, Baldet P, Joubès J, Dieuaide-Noubhani M, Just D, Chevalier C, Raymond P (2003) Physiological, biochemical and molecular analysis of sugar-starvation responses in tomato roots. J Exp Bot 54 1143–1151 [DOI] [PubMed] [Google Scholar]
- Dieuaide-Noubhani M, Canioni P, Raymond P (1997) Sugar-starvation-induced changes of carbon metabolism in excised maize root tips. Plant Physiol 115 1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004. a) Respiratory metabolism: glycolysis, the TCA cycle and the mitochondrial electron transport chain. Curr Opin Plant Biol 7 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004. b) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5 763–769 [DOI] [PubMed] [Google Scholar]
- Fitzgerald JN, Lehti-Shiu MD, Ingram PA, Deak KI, Biesiada T, Malamy JE (2006) Identification of quantitative trait loci that regulated Arabidopsis root system size and plasticity. Genetics 172 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Lea PJ (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58 2339–2358 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381 723–740 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A (2005) Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56 1469–1479 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Larher F (1997) Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal Biochem 251 153–157 [DOI] [PubMed] [Google Scholar]
- Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly M, Bothwell TH, Torrance JD, MacPhail AP, Derman DP, Bezwoda WR, Mills W, Charlton RW, Mayet F (1983) The effects of organic acids, phytates and polyphenols on the absorption of iron from vegetables. Br J Nutr 49 331–342 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449 1008–1013 [DOI] [PubMed] [Google Scholar]
- Imas P, Bar-Yosef B, Kafkafi U, Ganmore-Neumann R (1997) Phosphate induced carboxylate and proton release by tomato roots. Plant Soil 191 35–39 [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar TC, Ewing RP (1997) ROOTEDGE: software for measuring root length from desktop scanner images. Agron J 89 932–940 [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol 41 1030–1037 [DOI] [PubMed] [Google Scholar]
- Landschütze V, Müller-Röber B, Willmitzer L (1995) Mitochondrial citrate synthase from potato: predominant expression in mature leaves and young flower buds. Planta 196 756–764 [PubMed] [Google Scholar]
- Lemaitre T, Urbanczyk-Wochniak E, Flesch V, Bismuth E, Fernie AR, Hodges M (2007) NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol 144 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protocols 1 387–396 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Acevedo-Hernandez G, Ramirez-Chavez E, Molina-Torres J, Herrera-Estrella L (2006) Novel signals for plant development. Curr Opin Plant Biol 9 523–529 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramirez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella V, Herrera-Estrella L (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160 1–13 [DOI] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas Á, Bimbó A, Szabó L, Bánfalvi Z (2003) Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J 33 139–147 [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJJ, Kravchenko LV, Simons M (1999) Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol 1 439–446 [DOI] [PubMed] [Google Scholar]
- Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28 67–77 [DOI] [PubMed] [Google Scholar]
- Merckx R, Van Ginkel JH, Sinnaeve J, Cremers A (1986) Plant-induced changes in the rhizosphere of maize and wheat. I. Production and turnover of root-derived material in the rhizosphere of maize and wheat. Plant Soil 96 85–93 [Google Scholar]
- Millar AH, Leaver CJ (2000) The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett 481 117–121 [DOI] [PubMed] [Google Scholar]
- Müller-Röber B, Sonnewald U, Willmitzer L (1992) Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J 11 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Neumann G, Römheld V (1999) Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 211 121–130 [Google Scholar]
- Nguyen C, Todorovic C, Robin C, Christophe A, Guckert A (1999) Continuous monitoring of rhizosphere respiration after labelling of plant shoots with 14CO2. Plant Soil 212 189–199 [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007. a) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Sweetlove LJ, Fernie AR (2007. b) Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol Plant 129 45–56 [Google Scholar]
- Okamoto T, Tsurumi S, Shibasaki K, Obana Y, Takaji H, Oono Y, Rahman A (2008) Genetic dissection of hormonal responses in the roots of Arabidopsis grown under continuous mechanical impedance. Plant Physiol 146 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Moritz T, Cano-Delgado A, Harberd NP (1999) Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol 119 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet A, Raymond P (1983) Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Annu Rev Plant Physiol 34 199–224 [Google Scholar]
- Queval G, Noctor G (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363 58–69 [DOI] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI (2007) Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J 50 514–528 [DOI] [PubMed] [Google Scholar]
- Regierer B, Fernie AR, Springer F, Perez-Melis A, Leisse A, Koehl K, Willmitzer L, Geigenberger P, Kossmann J (2002) Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat Biotechnol 20 1256–1260 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development and cytokinin metabolism. Plant Cell 18 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Hirner B, Frommer WB (1993) Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell 5 1591–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Liu JL, Leisse A, Balbo I, Perez-Melis A, Willmitzer L, Fernie AR (2004) Kinetics of labelling of organic and amino acids in potato tubers by gas chromatography-mass spectrometry following incubation in 13C labelled isotopes. Plant J 39 668–679 [DOI] [PubMed] [Google Scholar]
- Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378 [DOI] [PubMed] [Google Scholar]
- Singer AC, Crowley DE, Thompson IP (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21 123–130 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP (2003) Normalization of cDNA microarray data. Methods 31 265–273 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Loukanina N, Ashihara H, Yeung EC, Thorpe TA (2003) Pyrimidine deoxyribonucleotide metabolism during maturation and germination of white spruce (Picea glauca) somatic embryos: metabolic fate of 14C-labelled cytidine, deoxycytidine and thymidine. Physiol Plant 118 499–506 [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR (2007) Reduced expression of succinyl CoA ligase can be compensated for by an upregulation of the β-amino-butyrate (GABA) shunt in illuminated tomato leaves. Plant Physiol 145 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Gibon Y, Frankel N, Wood CC, Zanor MI, Fernie AR, Carrari F (2005) Identification and characterisation of the alpha and beta subunits of succinyl CoA ligase of tomato. Plant Mol Biol 59 781–791 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39 792–796 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, Casimiro I, Peret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10 946–954 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Fait A, Nunes-Nesi A, Williams T, Fernie AR (2007) The mitochondrion: an integration point of cellular metabolism and signalling. Crit Rev Plant Sci 26 17–43 [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose 6-phosphate. Plant J 23 43–53 [DOI] [PubMed] [Google Scholar]
- Tcherkez G, Cornic G, Bligny R, Gout E, Ghashghaie J (2005) In vivo respiratory metabolism of illuminated leaves. Plant Physiol 138 1596–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomas S, Swarup R, Coates J, Swarap K, Laplaze L, Beemster GTS, Hedden P, Bhalerao R, Bennett MH (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10 625–628 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, et al (2006) Conversion of MapMan to allow the analysis of transcript data from solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60 773–792 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasula N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. . BMC Bioinformatics 7 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf L, Fromin N, Tomasi N, Aragno M, Martinoia E (2005) Secretion activity of white lupin's cluster roots influences bacterial abundance, function and community structure. Plant Soil 268 181–194 [Google Scholar]
- Wilkinson S, Davies WJ (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ 25 195–210 [DOI] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui R, Iketani S, Mikami T, Kubo T (2003) Antisense inhibition of mitochondrial pyruvate dehydrogenase E1α subunit in anther tapetum causes male sterility. Plant J 34 57–66 [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.