Abstract
Stem cells are proposed to segregate chromosomes asymmetrically during self-renewing divisions so that older (‘immortal’) DNA strands are retained in daughter stem cells whereas newly synthesized strands segregate to differentiating cells1–6. Stem cells are also proposed to retain DNA labels, such as 5-bromo-2-deoxyuridine (BrdU), either because they segregate chromosomes asymmetrically or because they divide slowly5,7–9. However, the purity of stem cells among BrdU-label-retaining cells has not been documented in any tissue, and the ‘immortal strand hypothesis’ has not been tested in a system with definitive stem cell markers. Here we tested these hypotheses in haematopoietic stem cells (HSCs), which can be highly purified using well characterized markers. We administered BrdU to newborn mice, mice treated with cyclophosphamide and granulocyte colony-stimulating factor, and normal adult mice for 4 to 10 days, followed by 70 days without BrdU. In each case, less than 6% of HSCs retained BrdU and less than 0.5% of all BrdU-retaining haematopoietic cells were HSCs, revealing that BrdU has poor specificity and poor sensitivity as an HSC marker. Sequential administration of 5-chloro-2-deoxyuridine and 5-iodo-2-deoxyuridine indicated that all HSCs segregate their chromosomes randomly. Division of individual HSCs in culture revealed no asymmetric segregation of the label. Thus, HSCs cannot be identified on the basis of BrdU-label retention and do not retain older DNA strands during division, indicating that these are not general properties of stem cells.
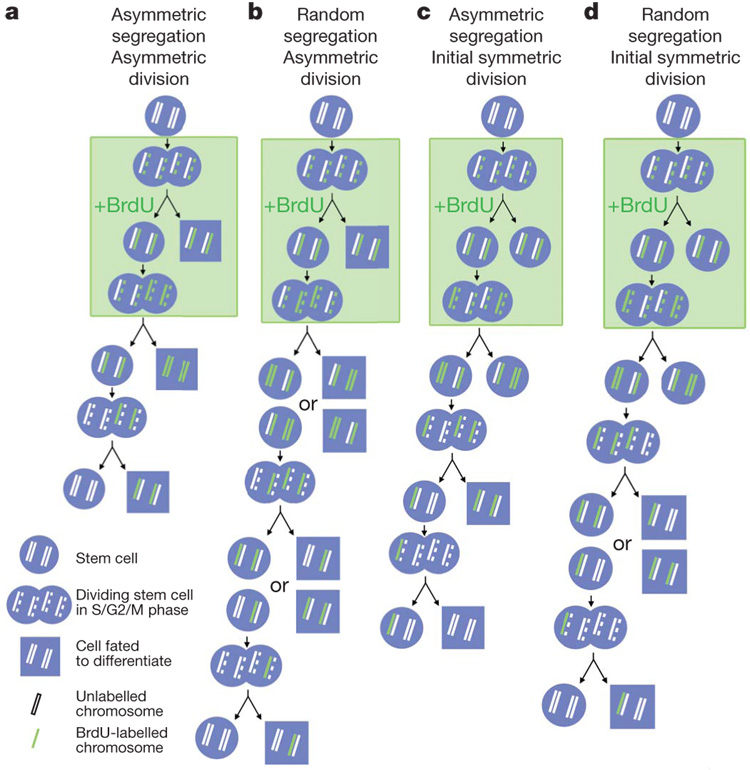
The immortal strand hypothesis was proposed as a mechanism by which stem cells could avoid accumulating mutations that arise during DNA replication2. Whereas most cells segregate their chromosomes randomly1,10, it was argued that adult stem cells in steady-state tissues might retain older DNA strands during asymmetric self-renewing divisions, segregating newly synthesized strands to daughter cells fated to differentiate (Fig. 1a). Evidence has supported this model in some epithelial stem cells1, neural stem cells3, mammary epithelial progenitors4 and muscle satellite cells5,6. A related idea is that adult stem cells in steady-state tissues might consistently retain DNAlabels. This could be because chromosomes segregate randomly but stem cells divide more infrequently than other cells (Fig. 1b), or alternatively because the older DNA strand is labelled and segregated asymmetrically (Fig. 1c). Tritiated thymidine8 or histone7 label-retaining cells from the hair follicle are enriched for epithelial stem cells, although the purity remains uncertain. Label-retaining cells have also been identified in the haematopoietic system9,11, in mammary epithelium12, in intestinal epithelium1,13 and in the heart14, but the purity of stem cells among these label-retaining cells has not been tested. As a result, it remains unclear whether label retention can consistently identify stem cells with specificity or sensitivity.
Figure 1. Contrasting predictions regarding stem cell labelling on the basis of the immortal strand model versus random chromosome segregation.
a, According to the immortal strand model2, stem cells divide asymmetrically under steady-state conditions and BrdU is incorporated into newly synthesized DNA strands that are asymmetrically segregated into differentiating daughter cells with each round of division, such that stem cells retain only the unlabelled older DNA strands. b, In contrast, if chromosomes segregate randomly, then BrdU-labelled chromosomes will be stochastically lost over multiple rounds of divisions. c, In the immortal strand model, if stem cells divide symmetrically then BrdU can be incorporated into DNA strands that become the ‘older’ strands once stem cells resume asymmetric division. Under these circumstances, the BrdU+ older strands would be retained indefinitely in stem cells. d, In contrast, if chromosome segregation is random then BrdU+ chromosomes are stochastically lost over time after BrdU is discontinued.
Under steady-state conditions in adult bone marrow, all HSCs divide regularly but infrequently15 to sustain haematopoiesis and to maintain nearly constant numbers of HSCs. As a result of this observation, as well as the finding that HSC divisions yield asymmetric outcomes in culture16, it has been proposed that adult HSCs divide asymmetrically16, although the rarity of HSCs in vivo and their relative quiescence has made it impossible to confirm this directly. Nonetheless, if BrdU-label retention and/or asymmetric chromosome segregation are general properties of adult stem cells, then either or both of these characteristics should be evident in HSCs, depending on experimental conditions.
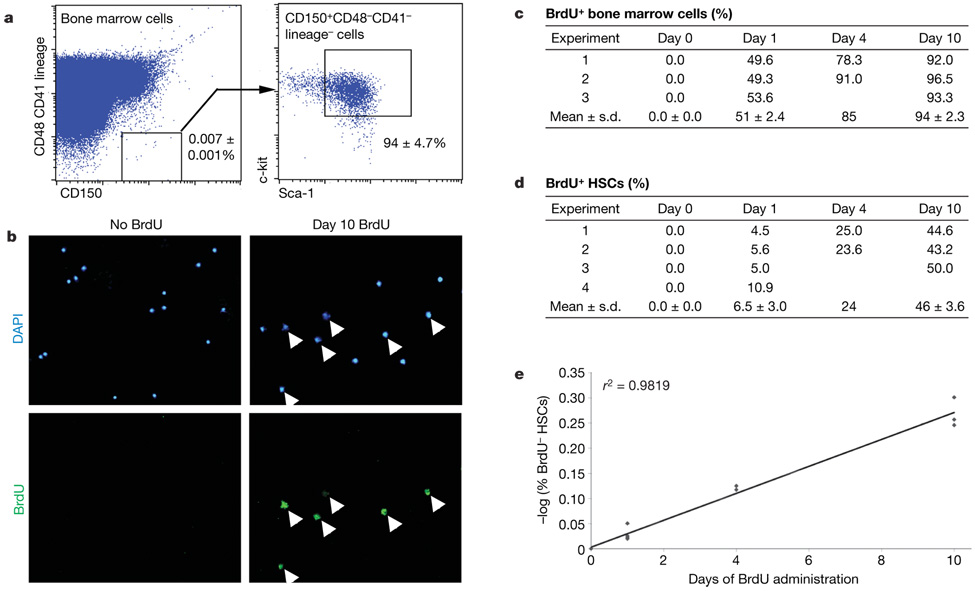
To test the rate at which HSCs enter the cell cycle we administered BrdU to mice for 1, 4 or 10 days, and then sorted HSCs onto microscope slides and stained with an anti-BrdU antibody. HSCs were sorted as CD150+CD48−CD41−lineage−Sca-1+c-kit+ cells (Fig. 2a). This population contains all of the detectable HSC activity in bone marrow, and 47% of single cells from this population give long-term multilineage reconstitution in irradiatedmice17. After 1–10 days of BrdU treatment, 51–94%of whole bone marrow cells and 6.5–45.9% of CD150+CD48− CD41−lineage−Sca-1+c-kit+ HSCs became BrdU+ (Fig. 2c, d). We calculated the rate at which HSCs entered the cell cycle15 to be 6.0%per day (Fig. 2e). Consistent with this, only 3.2% of CD150+CD48−CD41− lineage−Sca-1+c-kit+ cells were in S/G2/M phase of the cell cycle at any one time (Supplementary Fig. 3). These results are similar to a previous study that identified HSCs using different markers15.
Figure 2. Six per cent of HSCs stochastically enter the cell cycle each day.
a, HSCs can be isolated by flow cytometry as CD150+CD48−CD41−lineage−Sca-1+c-kit+ cells; these represent only 0.0066 ± 0.0003% (0.007% × 94%) of bone marrow cells but contain all detectable HSC activity and are very highly enriched for HSCs (nearly 50% of single cells give long-term multilineage reconstitution in irradiated mice17). b, BrdU incorporation into HSCs (arrowheads) is evaluated by immunofluorescence after sorting HSCs onto microscope slides (DAPI is a nuclear stain). c, d, The percentage of BrdU+ bone marrow cells (c) and CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs (d) after various periods of BrdU administration (3–4 independent experiments with 3–4 mice per experiment and 200–400 bone marrow cells or 100–400 HSCs counted, respectively, per mouse). Standard deviations are shown for means that are based on at least three independent experiments. e, The percentage of HSCs that enter the cell cycle each day (6%) can be derived by plotting the negative logarithm of the percentage of HSCs that were BrdU−over time15.
The linearity of BrdU incorporation over time (Fig. 2e) suggests that most HSCs divide at a similar rate. If a minority of HSCs divided more rapidly, consistently more than 6.0% of HSCs should have incorporated BrdU after one day; however, we did not observe this (Fig. 2d). If a minority of HSCs were more deeply quiescent than most other HSCs, these HSCs should remain BrdU-negative, even after long periods of BrdU treatment. This also has not been observed, because more than 99% of HSCs are labelled after 6 months of BrdU treatment 15. Therefore, there is no evidence for more rapidly dividing or more slowly dividing subsets of long-term self-renewing HSCs under steady-state conditions, although we cannot exclude the possibility that a minority of HSCs divide more slowly than 6.0% per day.
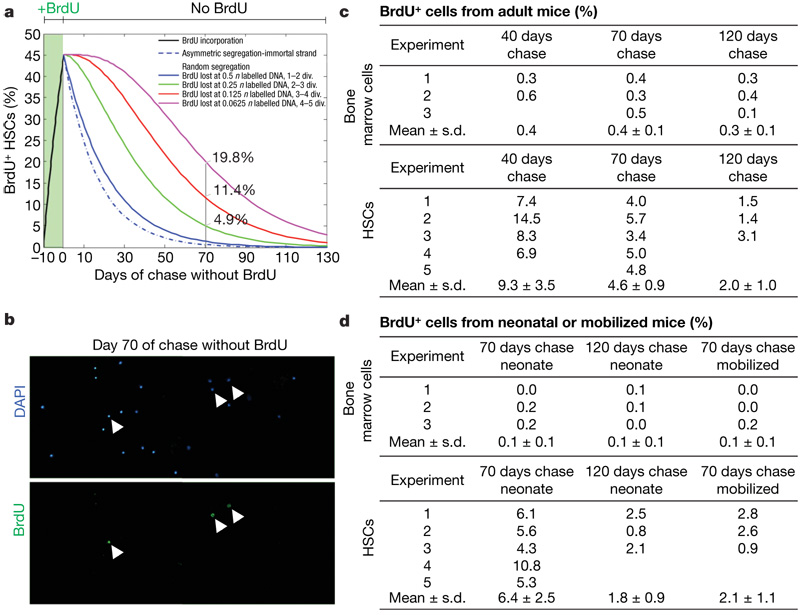
To evaluate retention of the BrdU label, we administered BrdU for 10 days, followed by 70 days without BrdU (a 70-day ‘chase’), like previous studies in the haematopoietic system9,11. Given that 6.0% of HSCs entered the cell cycle each day and 46% of HSCs were labelled after 10 days of BrdU treatment (Fig. 2d), we modelled the fraction of HSCs that would be expected to retain BrdU over time (Fig. 3a; see Methods for explanation).
Figure 3. Few HSCs retain BrdU, and most BrdU-retaining bone marrow cells are not HSCs.
a, A model is shown that predicts the fraction of HSCs that retain BrdU over time after administering BrdU for 10 days, depending on whether chromosomes segregate asymmetrically or randomly and on the threshold of BrdU that can be detected by immunofluorescence (0.5n, 0.25n, 0.125n or 0.0625n labelled DNA). b, CD150+CD48−CD41−lineage−Sca-1+ c-kit+ HSCs were sorted onto a microscope slide after 10 days BrdU administration and 70 days chase (without BrdU). Arrowheads identify BrdU+ cells. c, Shown is the frequency of BrdU+ bone marrow cells and HSCs after 10 days of BrdU administration and 40, 70 or 120 days of chase. Standard deviations are shown for means that are based on at least three independent experiments. d, Shown is the frequency of BrdU+ bone marrow cells and HSCs after 10 days BrdU administration to neonatal mice followed by 70 or 120 days of chase, or after 4 days BrdU administration to cyclophosphamide/G-CSF-mobilized mice followed by 70 days of chase. All data are based on 3–5 independent experiments with 3 mice per experiment and 400–700 bone marrow cells or 300–400 HSCs counted per mouse.
Under these experimental conditions, if HSCs follow the immortal strand model they should lose their BrdU label one division after BrdU is discontinued because the labelled chromosomes would be segregated to differentiating daughter cells (Fig. 1a); therefore, only 0.6% of HSCs would be expected to retain BrdU after the 70-day chase (Fig. 3a). If HSCs segregate chromosomes randomly, then BrdU would be lost stochastically over time and the fraction of BrdU+ HSCs after a 70-day chase would depend on the threshold of BrdU required for detection. If the threshold is equivalent to 0.5n labelled chromosomes (one-quarter of the genome), this level of BrdU dilution could be achieved in cells that had divided 1 or 2 times after BrdU was discontinued (depending on whether they had divided once or twice in the presence of BrdU), and only 1.4% of HSCs would be expected to retain BrdU after the 70-day chase (Fig. 3a). In contrast, if the threshold of detection is equivalent to 0.0625n labelled chromosomes, this could be achieved on average in cells that had divided 4 or 5 times after BrdU was discontinued, and 19.8% of HSCs would be expected to retain BrdU after the 70-day chase (Fig. 3a). Thus, these calculations predict that few (<20%) HSCs should retain BrdU after a 70-day chase, irrespective of how chromosomes are segregated.
To test this we administered BrdU for 10 days to adult mice and then stained whole bone marrow cells and CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs after 40, 70 and 120 days of chase. After 70 days of chase, 0.4 ± 0.1% (mean ± s.d.) of bone marrow cells and 4.6 ± 0.9% of HSCs were BrdU+ (Fig. 3c). This demonstrates that, as predicted (Fig. 3a), few HSCs retain BrdU. Moreover, only 2.0 ± 1.0% of HSCs were BrdU+ after 120 days of chase, demonstrating that the frequency of BrdU-retaining HSCs continues to decline over time rather than there being a deeply quiescent subset of HSCs that retains BrdU indefinitely. Although BrdU-label-retaining cells were enriched in HSCs by tenfold compared to whole bone marrow cells, the rarity of HSCs means that only 0.08% of BrdU+ bone marrow cells were HSCs (0.0066% × 4.6%/0.4%). BrdU-label retention is therefore a very insensitive and nonspecific marker of HSCs because most HSCs did not retain detectable BrdU, and only rare BrdU-label-retaining cells were HSCs. Very similar results were obtained when we used flow cytometry to detect BrdU incorporation (Supplementary Fig. 1) or when HSCs were isolated using different surface markers (c-kit+Flk-2−lineage−Sca-1+ cells; Supplementary Fig. 2). These data are most consistent with random chromosome segregation by HSCs and the failure to detect BrdU after approximately three divisions in the absence of BrdU (Fig. 3a).
According to the immortal strand model, stem cells can incorporate BrdU into DNA strands that become the ‘older’ strands during symmetric cell divisions, and these labelled DNA strands would be retained indefinitely by stem cells that resume asymmetric divisions (Fig. 1c). To test this, we administered BrdU to newborn mice for 10 days or to cyclophosphamide and granulocyte colony-stimulating factor (cyclophosphamide/G-CSF)-treated mice for 4 days. The absolute number of HSCs expands markedly in both newborn18 and cyclophosphamide/G-CSF-mobilized mice19 (indicating symmetric divisions), before stabilizing to steady-state levels as mice enter adulthood or after G-CSF is discontinued. After 10 days of BrdU treatment in neonatal mice, 93 ± 3.7% of bone marrow cells and 80 ± 11% of CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs were BrdU+. Seventy days later, 0.1% of bone marrow cells and 6.4 ± 2.5% of CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs were BrdU+ (Fig. 3d). After 4 days of BrdU treatment in cyclophosphamide/G-CSF mobilized mice, 94 ± 3% of CD150+CD48−CD41− lineage−Sca-1+c-kit+ HSCs were BrdU+. Seventy days later, 0.1% of bone marrow cells and 2.1 ± 1.1% of CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs were BrdU+ (Fig. 3d). Thus, even when BrdU was administered to symmetrically dividing HSCs, only 2–6% of HSCs retained the label and only 0.2–0.4% of BrdU-retaining bone marrow cells were HSCs. We were unable to identify any context in which BrdU-label retention identified HSCs with sensitivity or specificity, and none of these results was consistent with the immortal strand hypothesis.
To address the possibility that the HSCs in the above experiments might have continued to divide symmetrically after BrdU was discontinued, we also administered BrdU to mice from 20 to 29 days postnatally. HSCs are thought to transition from rapidly dividing cells that have a fetal phenotype to relatively quiescent cells that have an adult phenotype between 21 and 28 days postnatally18. We obtained similar results, with only 6.5 ± 1.1% of HSCs retaining BrdU after a 70-day chase. There was, therefore, no period during neonatal development when BrdU could be administered in a way that resulted in retention of BrdU within significant numbers of HSCs.
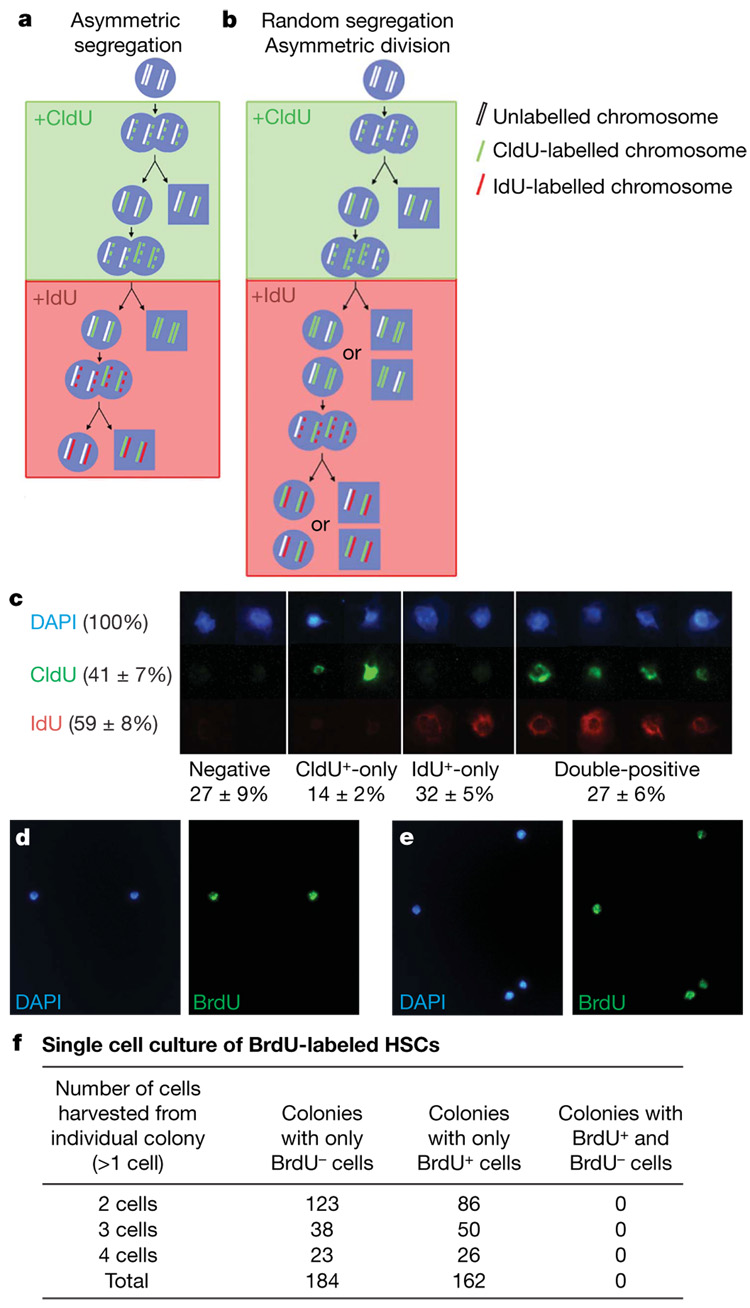
To test the immortal strand model directly, we treated mice with 5-chloro-2-deoxyuridine (CldU) for 10 days and then with 5-iodo-2-deoxyuridine (IdU) for 10 days. If HSCs segregate older and younger DNA strands asymmetrically, then HSCs should rarely incorporate both CldU and IdU under steady-state conditions because newly synthesized (labelled) DNA strands should be segregated to differentiating daughter cells after each division (Fig. 4a). In contrast, if HSCs segregate older and younger DNA strands randomly then CldU-labelled HSCs should have the same chance of incorporating IdU as unlabelled cells, and approximately 25% (50% × 50%) of HSCs should be double-labelled (Fig. 4b).
Figure 4. HSCs segregate chromosomes randomly in vivo and in vitro.
a, By the immortal strand model, stem cells sequentially exposed to CldU (for 10 days) and then IdU (for 10 days) would not incorporate both labels, with the exception of rare cells in the S/G2/M phase of their first division after switching from CldU into IdU (expected frequency <3%). b, In contrast, if chromosome segregation is random then CldU+ stem cells would have the same probability of incorporating IdU as unlabelled cells (expected frequency of CldU+IdU+ HSCs is ~25%). c, CldU was administered to mice for 10 days followed by IdU for 10–11 days, and CD150+CD48−CD41−lineage−Sca-1+ c-kit+ HSCs were stained. Examples of HSCs that incorporated neither label, only CldU, only IdU or both labels are shown (mean ± sd; data are based on two independent experiments with 2 or 3 mice per experiment and 100–250HSCs per mouse).HSCs fromBrdU-treated mice divided once (d)or twice (e) in culture to form daughter cells. f, The progeny of these HSCs were either all BrdU+ or all BrdU−. We detected no clones in which label was asymmetrically segregated to a subset of daughter cells.
After 10 days of CldU followed by 10–11 days of IdU, we observed that 14% of HSCs incorporated only CldU, 32% incorporated only IdU and 27% incorporated both CldU and IdU (Fig. 4c and Supplementary Fig. 4). The frequency of CldU+IdU+ cells (27%) was therefore similar to the product of the frequencies of total CldU+ cells and total IdU+ cells (41% × 59% = 24%), indicating that CldU+ cells had a similar probability of incorporating IdU as the other cells. We repeated this experiment by administering mice with CldU for 60 days followed by IdU for 15 days, and found the frequency of CldU+IdU+ cells (63%) was again similar to the product of the frequencies of total CldU+ cells and total IdU+ cells (73% × 84% = 61%). These results were not significantly affected by a slow clearance of CldU from mice, because CldU was cleared in less than 1 day after being discontinued from the drinking water (Supplementary Fig. 5). These observations directly contradict a key prediction made by the immortal strand hypothesis, but are as would be expected by random chromosome segregation.
The foregoing experiments left the formal possibility that if HSCs divide by a combination of symmetric and asymmetric divisions in vivo we might underestimate the frequency of HSCs that retain older DNA strands. To address this we examined the division of individual HSCs in culture that were isolated from mice treated for 10 days with BrdU. Single CD150+CD48−CD41−lineage−Sca-1+c-kit+ HSCs were sorted into cultures under conditions in which half of HSC divisions give asymmetric outcomes (daughter cells with different developmental potentials)16. After 2–3 days of culture we observed a total of 346 colonies in which HSCs had divided once (2 daughter cells) or twice (3 or 4 daughter cells). Either all daughter cells were BrdU+ (162 colonies, 46%) or all daughter cells were BrdU− (184 colonies; Fig. 4f), as would be expected by random chromosome segregation given that 46% of HSCs incorporate BrdU in vivo after 10 days (Fig. 2d). We observed no colonies containing a mixture of BrdU+ and BrdU− cells after one or two rounds of division. Thus, these in vitro experiments on individual HSCs failed to detect any asymmetric segregation of labelled chromosomes.
Our results were not confounded by the effects of BrdU, CldU or IdU on HSC proliferation, survival or DNA repair. The cell cycle status (Supplementary Fig. 3) and frequency (data not shown) of HSCs was not affected by these treatments. BrdU, CldU and IdU incorporation from DNA repair was not detectable (Supplementary Fig. 3).
Our results indicate that BrdU-label retention is neither a sensitive nor a specific marker of HSCs. Nonetheless BrdU-label retention could be a better marker of stem cells in other tissues. Moreover, histone–green fluorescent protein retention7 may do a better job of marking stem cells, including HSCs, because it can be selectively expressed in subsets of progenitors and may be retained with different kinetics than BrdU. Our data demonstrate the need to test the sensitivity and specificity of BrdU and other label-retention markers before assuming they mark stem cells with fidelity.
Our data also demonstrate that the immortal strand model2 does not apply to HSCs and cannot be considered a general model of stem cell division. Our data do not address whether stem cells from other tissues asymmetrically segregate chromosomes or whether HSCs segregate a limited number of chromosomes asymmetrically10. Nonetheless, asymmetric chromosome segregation cannot be a mechanism by which HSCs avoid accumulating mutations over time.
METHODS SUMMARY
BrdU, CldU and IdU administration
All experiments used C57BL/Ka-CD45.2:Thy-1.1 mice. For experiments in adult mice, BrdU (Sigma) was administered when the mice were 8–10 weeks of age. Mice were given an intraperitoneal injection of 100mg BrdU per kg body mass in Dulbecco’s phosphate buffered saline (DPBS; Gibco) and were maintained on 1 mg ml−1 BrdU in the drinking water for 1–10 days. Amber bottles containing BrdU water were changed every 1–3 days. For retention studies, BrdU water was replaced with regular water and the mice were maintained for 40–120 days before analysis.
BrdU injections into neonatal mice were performed as described20. Beginning within 3 days after birth, neonatal mice were injected subcutaneously with 50mg BrdU per kg body weight twice daily for 10 days. Mice were weighed every 2 days and the dose of BrdU was adjusted.
To assess BrdU retention after cytokine mobilization, adult mice were injected with cyclophosphamide (200mg kg−1; Bristol-Myers Squibb) and then on each of the 4 subsequent days they were injected with 250 µg kg−1 day−1 of human G-CSF (Amgen)19. On the fourth day of G-CSF injection, a single intraperitoneal injection of 100 mg BrdU per kg body mass was given and the mice were put on 1 mg ml−1 BrdU water for 4 additional days. The mice were then returned to regular water for 70 days before analysis.
For CldU and IdU experiments, mice were given an intraperitoneal injection of 100 mg CldU per kg body mass in DPBS and were maintained on drinking water containing 1 mg ml−1 CldU (Sigma) for 10 days. Mice were then given an intraperitoneal injection of 100 mg IdU per kg body mass in DPBS, and were switched to drinking water containing 1 mg ml−1 IdU (Sigma) for 10–11 days before being killed.
For details related to the flow cytometric isolation of HSCs and CldU, IdU and BrdU staining, see Methods.
BrdU segregation in cultured HSCs
Single CD150+CD48−CD41−lineage−Sca-1+ c-kit+ HSCs were sorted from BrdU-treated mice into a V-bottom 96-well plate containing Stempro-34 medium (Invitrogen) supplemented with 2mM l-glutamine, 50 µM 2-mercaptoethanol (Sigma), murine IL-3 (10 ng ml−1), murine SCF (100 ng ml−1) and murine Tpo (100 ng ml−1; all cytokines were obtained from R&D Systems) with 10% charcoal absorbed fetal bovine serum (Cocalico Biologicals Inc.), and were cultured for 2–3 days in low-oxygen chambers21. For analysis, plates were centrifuged at 500g for 10 min; cells from each colony were then pipetted onto individual wells of Teflon-printed glass slides and were allowed to dry overnight before staining for BrdU and 4,6-diamidino-2-phenylindole (DAPI).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, the National Institute on Aging (NIH), and the US Army Research Laboratory/Office. Flow cytometry was partially supported by the UM-Comprehensive Cancer Centre and the UM-Multipurpose Arthritis Centre. Antibody production was partially supported by the Rheumatic Core Disease Centre. M.J.K. was supported by a University of Michigan Cancer Biology Training Grant. The authors thank D. Adams and M. White for flow cytometry and E. Smith (Hybridoma Core Facility) for antibody production.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 3.Karpowicz P, et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J. Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 5.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 6.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 10.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311:1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 11.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Welm BE, et al. Sca-1+ cells in the mouse mammary gland represent an enriched progenitor cell population. Dev. Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 13.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 14.Urbanek K, et al. Stem cell niches in the adult mouse heart. Proc. Natl Acad. Sci. USA. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takano H, Ema H, Sudo K, Nakauchi H. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J. Exp. Med. 2004;199:295–302. doi: 10.1084/jem.20030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Bowie MB, et al. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Invest. 2006;116:2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison SJ, Wright D, Weissman IL. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl Acad. Sci. USA. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 21.Morrison SJ, et al. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J. Neurosci. 2000;20:7370–7376. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.