Abstract
Leaf organogenesis occurs within the peripheral zone of the shoot apical meristem (SAM). It has been known that several members of the class-1 KNOTTED1-like homeobox (KNOX) genes are expressed in the SAM, and their expression must be prevented during leaf primordium initiation and subsequent leaf development. A number of regulators that repress class-1 KNOX genes have been identified, and characterizations of these regulators greatly improved our knowledge of the genetic basis of leaf organogenesis. We have recently reported that the proteolytic function of the Arabidopsis 26S proteasome is involved in specifying leaf adaxial identity during leaf development, by characterizations of mutants defective in genes encoding several 26S proteasome subunits. Here we demonstrate that in addition to the role in leaf polarity establishment, the 26S proteasome also participates in repression of class-1 KNOX genes during leaf development. We show that loss of functions in RPN8a and RPT2a, two 26S proteasome subunit genes, resulted in leaves that produce ectopic outgrowths on the abaxial side of blades. These outgrowths were accompanied by the ectopic expression of several class-1 KNOX genes. These results indicate that the 26S proteasome is important in repressing class-1 KNOX genes and its function may be required until later leaf developmental stages.
Key Words: 26S proteasome, AE3/RPN8a, class-I KNOX genes, leaf development
Class-1 KNOX genes are members belonging to the super homeobox gene family, and are found in species throughout the plant kingdom.1 The class-1 KNOX genes are expressed in the SAM for maintaining meristematic identity, whereas are downregulated at the position where the leaf initiates. It has been known that persistent repression of class-1 KNOX genes is required for subsequent leaf development.2,3 In Arabidopsis, class-1 KNOX genes consists of four members: SHOOT MERISTEMLESS (STM), BREVIPEDICELLUS (BP), KNAT2 and KNAT6. Ectopic expression of BP led to meristem- and organ-like structures forming on the leaf, or resulted in leaves with other phenotypic abnormalities.4–7 To date, a number of genes have been found to play roles in repressing class-1 KNOX genes during leaf development. These include ASYMMETRIC LEAVES1/2,5,8,9 HIRA,10 YABBYs,6 BLADE-ON-PETIOLE1/2,7,11,12 SERRATE (SE) and PICKLE (PKL),5 FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and CURLY LEAF (CLF) complex,13 and even posttranscriptional gene silencing components RNA-DEPENDENT RNA POLYMERASE (RDR6)14 and ARGONAUTE1 (AGO1).15
The regulated protein degradation by the ubiquitin/26S proteasome system is crucial for a wide range of developmental and physiological processes in eukaryotes.16 Recently, we reported a role of the 26S proteasome in Arabidopsis leaf polarity formation.17 We demonstrate here that during leaf development the 26S proteasome is also involved in repression of class-1 KNOX genes. In comparison to the wild-type leaves (Fig. 1A), the ae3-1 mutant, which carries a lesion in the 26S proteasome RPN8a subunit,17 produced narrow rosette leaves (Fig. 1B and C). In some rosette leaves and most cauline leaves, ectopic outgrowths formed on the abaxial side of leaf blade, associating with the midrib in the distal region (Fig. 1D and 1E). Some of these outgrowths could eventually form leaflets (Fig. 1E).17 These abnormal leaf phenotypes can be observed in some other 26S proteasome subunit mutants, such as hlr-2, which harbors a defective RPT2a (Fig. 1F). These ectopic outgrowths indicate that ae3-1 and hlr-2 mutations may restore the meristematic capacity in the leaf.
Figure 1.
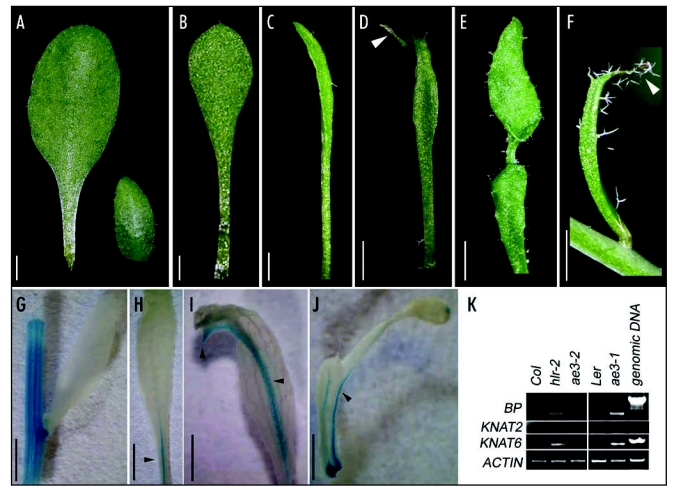
Altered leaf phenotypes and ectopic expression of class-1 KNOX genes in ae3-1 and hlr-2 plants. (A) A rosette leaf (left) and a cauline leaf (right) from a wild-type Landsberg erecta plant. (B–E) ae3-1 leaves. Most rosette leaves of ae3-1 became long and narrow (B), and some of them were very narrow (C). Ectopic outgrowths were formed on abaxial side of some rosette and cauline leaves, associating with the midrib in the distal region of blades. Arrowhead points to an outgrowth appearing on a rosette leaf (D). Some of the outgrowths extend their growth to form leaflet on cauline leaves (E). (F) An hlr-2 cauline leaf with an ectopic outgrowth (arrowhead). (G–J) Analyses of GUS activity in leaves carrying a BP::GUS fusion in wild type (G) and ae3-1 mutant plants (H–J). Note that leaves with outgrowths usually showed stronger GUS staining. (K) RT-PCR analyses of BP, KNAT2 and KNAT6 expression in ae3-1, ae3-2 and hlr-2 leaves. Note that ae3-1 (Ler) and hlr-2 (Col) leaves used in the RT-PCR experiment had ectopic outgrowths, whereas the ae3-2 (Col) had not such structures. Bars = 2 mm in (A–F), and 5 mm in (G–J).
To examine whether these outgrowths were also accompanied by the ectopic expression of class-1 KNOX genes, we analyzed wild-type and mutant plants carrying a BP::GUS fusion. In wild-type plants (Fig. 1G), GUS staining was detected in the stem but not in the leaf blade. By contrast, most ae3-1 leaves showed GUS staining in the midrib, although the intensity of GUS staining differed among mutant leaves (Fig. 1H–J). Interestingly, leaves with the outgrowth usually showed stronger GUS staining (Fig. 1I and J) than those that had no ectopic outgrowth (Fig. 1H). We also performed reverse transcription-polymerase chain reaction (RT-PCR) to analyze BP, KNAT2 and KNAT6 expression in wild-type and mutant leaves. Strikingly, BP, KNAT2 and KNAT6 transcripts were present in ae3-1 and hlr-2 leaves that carried outgrowths, whereas were absent in wild-type leaves or nearly undetectable in the mutant leaves that had no ectopic outgrowth (Fig. 1K). These results suggest that the outgrowths on the ae3-1 and hlr-2 leaves may be caused by the ectopic expression of class-1 KNOX genes.
In Arabidopsis, genes that repress class-1 KNOX members can be grouped into two general types, based on the ectopic expression of KNOX genes in the single gene mutation or in the combined gene mutations.15 The first category (type I) includes AS1/2, BOP1/2, FILAMENTOUS FLOWER, YABBY3, AGO1, FIE and CLF. The mis-expression of all or some class I KNOX genes can be detected in the leaves of plants with these gene mutations. The second category (type II) comprises SE, PKL and RDR6. The class I KNOX genes are normally repressed in the se, pkl, rdr6 single mutant plant leaves, but are expressed more robustly in the se as1(2), pkl as1(2) and rdr6 as1(2) double mutant leaves.5,14 Our results showed that the 26S proteasome subunit genes belong to the type I KNOX repressor category.
Taken together, our data reveal that the 26S protein degradation machinery is important in leaf developmental regulation, not only for leaf polarity establishment but also for class-1 KNOX gene repression. It would be of great interest to identify in the future the targets of the 26S proteasome, and to uncover whether the targets share functions in regulating both KNOX and polarity genes.
Acknowledgements
This work is supported by grants 30630041, 30421001, KSCX2-YW-N-016 and 04JC1-4077 to H.H.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3645
References
- 1.Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. The role of knox genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- 2.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 3.Tsiantis M, Hay A. Comparative plant development: the time of the leaf? Nat Rev Genet. 2003;4:169–180. doi: 10.1038/nrg1002. [DOI] [PubMed] [Google Scholar]
- 4.Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- 6.Kumaran MK, Bowman JL, Sundaresan V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell. 2002;14:2761–2770. doi: 10.1105/tpc.004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha CM, Kim GT, Kim BC, Jun JH, Soh MS, Ueno Y, Machida Y, Tsukaya H, Nam HG. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- 8.Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene Asymmetric Leaves2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Xu Y, Dong A, Sun Y, Pi L, Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- 10.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1361–1370. doi: 10.1093/pcp/pch201. [DOI] [PubMed] [Google Scholar]
- 12.Norberg M, Holmlund M, Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005;132:2203–2213. doi: 10.1242/dev.01815. [DOI] [PubMed] [Google Scholar]
- 13.Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–719. doi: 10.1111/j.1365-313x.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Huang W, Wang H, Cai R, Xu Y, Huang H. Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol Biol. 2006;61:63–78. doi: 10.1007/s11103-005-5992-7. [DOI] [PubMed] [Google Scholar]
- 16.Smalle J, Vierstra RD. The ubiquitin 26s proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The Proteolytic Function of the Arabidopsis 26S Proteasome Is Required for Specifying Leaf Adaxial Identity. Plant Cell. 2006;18:2479–2492. doi: 10.1105/tpc.106.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]


