Abstract
Mitogen activated protein kinases (MAPKs) are prevalent signal transduction proteins in eukaryotes, and play multiple and important roles by responding to a variety of stimuli. Numerous papers provided evidence for extensive use of these modules in plants, and some recently emerging data might seem difficult to reconcile with previously reported studies. Here, we illustrate the difficulties and current challenges of studying plant MAPKs by discussing published studies on pathways comprising MEKK1, MKK1 and MPK4.
Key Words: mitogen activated protein kinase, signaling, network, plant research methods
Mitogen activated protein kinases (MAPKs) are prevalent signal transduction proteins in eukaryotes, and play multiple and important roles by responding to a variety of stimuli. Numerous papers provided evidence for extensive use of these modules in plants, and some recently emerging data might seem difficult to reconcile with previously reported studies. Here, we illustrate the difficulties and current challenges of studying plant MAPKs by discussing published studies on pathways comprising MEKK1, MKK1 and MPK4 (Fig. 1).
Figure 1.
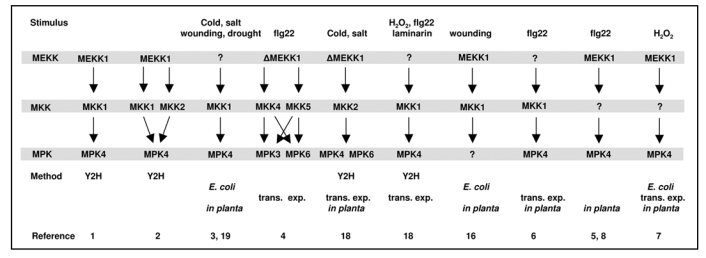
Research data on signal transduction pathways comprising MEKK1, MKK1 and MPK4. Abbreviations of methods: Y2H, yeast two-hybrid analysis; E. coli, interaction studies with bacterially overexpressed proteins; in planta, studies with protein complexes purified from plants and phenotypic analysis of mutants; trans. exp., interaction studies with proteins overexpressed in protoplasts.
The MEKK1-MKK1-MPK4 cascade was the first complete putative MAPK module delineated in plants using yeast two-hybrid (Y2H) system,1 and subsequent in vitro data confirmed the existence of this complex.2,3 Later on, a contradictory paper based on transient expression of wild-type, gain of function and dominant negative constructs described MEKK1 as the upstream kinase of MKK4/5-MPK3/6 in a pathway downstream of the flagellin receptor FLS2.4 In year 2006, four other laboratories presented in planta data relating to a MEKK1-MKK1-MPK4 module using T-DNA insertion mutants of MEKK1 and MKK1, and measuring MPK3, MPK4 and MPK6 activities in these plants in response to pathogenic elicitors and H2O2.5–8 To reconcile these seemingly opposing results, we need to consider the origin of these data and the methodological pitfalls of the different approaches. Limitations of Y2H are known and taken into account when presenting data based on this system,9 but what of other methods currently used in the plant signaling field?
Transient expression in protoplasts has now become a high-throughput and effective way to identify plant protein interactions and activities of pathways.10 The main benefit of this system is that proteins of interest are allowed to find interacting partners in their natural environment, i.e., inside plant cells. Furthermore, protoplasts possess their own signal transduction components; hence, biological responses to different stimuli can be studied without need for exogenously applied components. However, some drawbacks of overexpression are shared with Y2H, for ectopic expression can result in artificial protein interactions and thus can lead to inappropriate activation of pathways and physiological responses. Therefore, results obtained using transient expression systems -like Y2H data- should be confirmed by other methods.
To ensure higher or constitutive kinase activity, MAPKKKs (MEKKs) lacking their N-terminal regulatory domain are often used in signal transduction studies. Although these forms were shown to retain some specificity to selectively activate downstream components and thus provided important insights into plant MAPK signaling, generated data must be handled with caution. The early finding of a direct interaction between MPK4 and MEKK1 suggested a possible scaffolding function for MEKK1.2 This hypothesis gained further support when MEKK1 was shown to bind in vitro specifically to MPK4, MPK5 and MPK13.7 Similarly, the closest homologue of MEKK1 in alfalfa, MsOMTK1, can physically interact with the MAPK MsMKK3 and therefore was also proposed to serve as a scaffold. A truncated version of MsOMTK1 lacking its N-terminal domain activated MsMKK3 stronger than the wild-type kinase, but showed reduced MsMKK3 binding and cell death inducing capacity.11 These findings are actually not unexpected, since in yeast and mammals scaffolding proteins are central to the organization of signaling complexes, and MAPK components themselves, such as Pbs2 in yeast or MEKK2 in human, are known to function as scaffolds.12 13 Deleting domains that carry scaffolding properties might disrupt the functionality of these kinases and result in the activation of inappropriate signaling cascades.
To best assess in vivo mechanistics of MAPK modules, we need to set up model systems where these modules work, i.e., in plants. As expected, this is the most time-consuming and technically most challenging approach; additionally, generated data can be misleading.14 In planta studies are difficult to interpret since constitutively altered protein levels lead to long-term perturbations and also might result in different effects in distinct plant tissues. Two laboratories analyzed changes in MAPK activity upon flg22 or H2O2 treatment in mekk1 knockout plants, and both observed reduced MPK4 activity.5,7 The effect on MPK3 and MPK6 activities however, were not as clear, since their activity was found to be elevated in seedlings, but slightly reduced in roots. This might result from differing stress treatments, but it could be equally important that, depending on their cellular environment, the same kinases might be involved in several protein complexes and fulfill different functions. We discussed previously problems relating to the analysis of knockout mutants in case of redundancy and/or overlapping functions within a family of genes.6
In our publication, we faced the challenge to reconcile all previously published data on the existence of MEKK1-MKK1/2-MPK4 and MEKK1-MKK4/5-MPK3/6 pathways with our data on mkk1 knockout, that is hypersensitive to Pseudomonas and displays lower MPK4, MPK3 and MPK6 activity than wild-type in response to flagellin. An additional complication to consider is that MKK4/5 and MPK3/6 are positive regulators of pathogen responses while MPK4 is a negative regulator.4,15 According to our models, MPK4 and MKK1, besides having a direct effect on pathogen responses, regulate the capacity of MPK3 and MPK6 to bind MEKK1 including complex. In one scenario, we proposed that MEKK1 has a higher affinity for MKK1 in complex with MPK4, therefore MKK4/5 can only be activated after release of the MKK1-MPK4 complex (sequential activation model). We now know that MEKK1 is not required for MPK3/6 activation, and that removing it can actually hyperactivate the MPK3/6 pathway.5 Thus, the second scenario is more likely, where MKK4/5 is trapped by MEKK1 in a non-functional complex and released upon stimulation to be activated by a distinct upstream MAPKKK (trapping model). This model is further supported by in vivo data showing that N-terminally truncated MEKK1 specifically phosphorylates MKK1 and MKK2, but neither MKK4 nor MKK5.16 Considering the dispensability of MEKK1 for MPK3/6 activation, an alternative hypothesis can be proposed to explain the interconnected MPK3, MPK4 and MPK6 activities. MPK3/6 might not be activated or bound by a MEKK1-containing complex, but rather could be indirectly regulated by the MEKK1-MKK1-MPK4 module, e.g., through the activation of a phosphatase. Understanding how these separate pathways communicate will require the isolation of the relevant complexes. The upstream MAPKKK of MKK4/5 and MPK3/6 in the last two scenarios also remains to be identified.
How could we answer the above questions and more generally elucidate plant signal transduction pathways? Signaling modules never exist in isolation but are part of intricate networks and operate on the basis of combinatorial interactions. Description of complete protein linkage maps -interactomes- calls for robust methods like high throughput Y2H assays or identification of in vivo complexes by mass spectrometry. However, protein-protein interactions are only one aspect of the regulation of signaling modules. A historical overview of cell signaling studies discussed three other crucial themes related to MAPK regulation.17 First, scaffolding proteins organize relevant protein complexes; second, localization can dictate the biological response; third, duration of MAPK activity influences the final outcome of triggered pathway. To understand the in vivo functioning of signaling pathways, we need to turn to more sophisticated methods—e.g., stable and conditional mutants, inducible and tissue specific gene expression, protein localization. Studying the dynamic changes upon perturbation at a system-wide level (variations in transcriptome, proteome, phosphoproteome and metabolome activities) should allow in the future to gain deeper insights into how signaling pathways are flexibly used to channel, process and integrate cellular information. This arduous task is beyond the capacity of a single laboratory. A joint effort from multiple scientists will produce relevant yet potentially contradictory results, and the challenge will be to position all data into coherent signaling networks.
Abbreviations
- MAPK/MPK
mitogen activated protein kinase
- Y2H
yeast two-hybrid
- MKK
MAPK kinase
- MEKK
MKK kinase
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3885
References
- 1.Mizoguchi T, Ichimura K, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 1998;437:56–60. doi: 10.1016/s0014-5793(98)01197-1. [DOI] [PubMed] [Google Scholar]
- 2.Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K. Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun. 1998;253:532–543. doi: 10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- 3.Huang YF, Li H, Gupta R, Morris PC, Luan S, Kieber JJ. ATMPK4, an arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiology. 2000;122:1301–1310. doi: 10.1104/pp.122.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 5.Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem. 2006;281:36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- 6.Meszaros T, Helfer A, Hatzimasoura E, Magyar Z, Serazetdinova L, Rios G, Bardoczy V, Teige M, Koncz C, Peck S, Bogre L. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006;48:485–498. doi: 10.1111/j.1365-313X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- 7.Nakagami H, Soukupova H, Schikora A, Arsky V, Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem. 2006 doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 8.Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2006 doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causier B, Davies B. Analysing protein-protein interactions with the yeast two-hybrid system. Plant Molecular Biology. 2002;50:855–870. doi: 10.1023/a:1021214007897. [DOI] [PubMed] [Google Scholar]
- 10.Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagami H, Kiegerl S, Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J Biol Chem. 2004;279:26959–26966. doi: 10.1074/jbc.M312662200. [DOI] [PubMed] [Google Scholar]
- 12.O'Rourke SM, Herskowitz I, O'Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Yang J, Xia Y, Karin M, Su B. Synergistic interaction of MEK kinase 2, c-Jun N-terminal kinase (JNK) kinase 2, and JNK1 results in efficient and specific JNK1 activation. Mol Cell Biol. 2000;20:2334–2342. doi: 10.1128/mcb.20.7.2334-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bent AF. Plant mitogen-activated protein kinase cascades: Negative regulatory roles turn out positive. P Natl Acad Sci USA. 2001;98:784–786. doi: 10.1073/pnas.98.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 16.Hadiarto T, Nanmori T, Matsuoka D, Iwasaki T, Sato K, Fukami Y, Azuma T, Yasuda T. Activation of Arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1-AtMEK1 pathway by wounding. Planta. 2006;223:708–713. doi: 10.1007/s00425-005-0126-7. [DOI] [PubMed] [Google Scholar]
- 17.Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 18.Teige M, Scheikl E, Eulgem T, Doczi F, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Molecular Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T. Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 2002;29:637–647. doi: 10.1046/j.0960-7412.2001.01246.x. [DOI] [PubMed] [Google Scholar]


