Abstract
Due to their wall-associated pectin metabolism, growing plant cells emit significant amounts of the one-carbon alcohol methanol. Pink-pigmented microbes of the genus Methylobacterium that colonize the surfaces of leaves (epiphytes) are capable of growth on this volatile C1-compound as sole source of carbon and energy. In this article the results of experiments with germ-free (gnotobiotic) sporophytes of angiosperms (sunflower, maize) and gametophytes of bryophytes (a moss and two liverwort species) are summarized. The data show that methylobacteria do not stimulate the growth of these angiosperms, but organ development in moss protonemata and in thalli of liverworts is considerably enhanced. Since methylobacteria produce and secrete cytokinins and auxin, a model of plant-microbe-interaction (symbiosis) is proposed in which the methanol-consuming bacteria are viewed as coevolved partners of the gametophyte that determine its growth, survival and reproduction (fitness). This symbiosis is restricted to the haploid cells of moisture-dependent “living fossil” plants; it does not apply to the diploid sporophytes of higher embryophytes, which are fully adapted to life on land and apparently produce sufficient amounts of endogenous phytohormones.
Key Words: epiphytes, coevolution, symbiosis, methylobacteria, phytohormones, phyllosphere, plant-microbe interaction
Introduction
In the last December Issue 06 of the journal Nature it was reported that the intestinal bacteria (“gut flora”) in obese humans and mice differ from those in lean individuals of the same population.1 This high-impact Cover story entitled “The role of gut microbes in obesity” was based on experiments with germ-free wild-type mouse recipients (gnotobiotics). The authors concluded that the gut microbiota may be an additional contributing factor to the pathophysiology of obesity in mice and humans.1 The widely used term “gut flora” perpetuates an outdated classification of the bacteria as plants; it has been replaced by “microbiota”, which is defined as the sum of all prokaryotic microbes (Bacteria, Archaea) that inhabit a specific ecological niche.
A number of reports indicate that the beneficial bacteria in the human gut are coevolved microbial partners (intestinal symbionts) of their multicellular eukaryotic host.2 Corresponding experiments with axenic plants as host organisms are described in this article. First, I provide some background information on phyllosphere microbiology. Then, I summarize recent studies with germ-free (gnotobiotic) land plants (embryophytes) that were inoculated with certain bacteria. Based on these results I propose a specific plant-microbe interaction with reference to a recent original article3 on this topic.
Plant-Associated Bacteria in the Phyllosphere
The stem and plant leaf surface, or above-ground phytosphere, is a suitable habitat for a variety of microorganisms (Fig. 1A). Under natural conditions, communities of bacteria and fungi, which represent the most commonly detected residents on the outer cuticular layer of the epidermal cells, can vary considerably from one organ to another. The variability in bacterial population densities may in part be attributable to the harsh conditions on the surfaces of aerial plant organs such as leaves; this so-called “phyllosphere” is exposed to steadily changing environmental factors (wind, rain, sunlight etc.). The surface of the root (“rhizosphere”), however, represents a less variable environment for microbes. This habitat is denoted as the below-ground phytosphere.4
Figure 1.
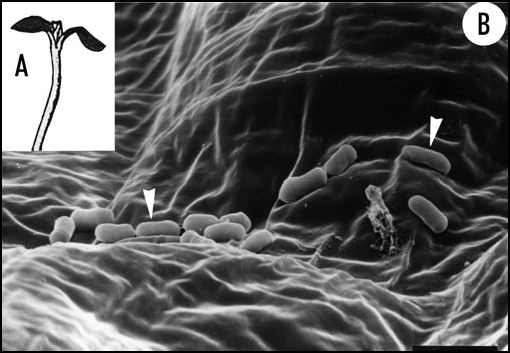
Epiphytic bacteria on the surface of the epidermal cells of a higher plant. Five-day-old light-grown seedling of sunflower (Helianthus annuus) that was raised under non-sterile standard conditions (A). A scanning electron micrograph of a cotyledon (lower epidermis) reveals the presence of numerous bacteria (arrow heads) (B). Since the host organism is a healthy, rapidly growing sporophyte these microbes must be interpreted as non-pathogenic commensals (or symbionts) of the plant. Bar = 2 µm.
Epiphytic bacteria, or “epiphytes” (Fig. 1B), can be defined as microbes that are surface-dwelling and free-living. They are usually capable of growing (multiplying) on the surface of their eukaryotic host organism, although the undamaged phyllosphere (leaf surface) is poor in organic nutrients. Insect feeding, frost damage or other agents that cause mechanical wounding of single plant cells can induce the leakage of intracellular sap that is rich in organic nutrients (sugars etc.). However, some reports document that even undamaged epidermal cells constantly leak minute amounts of metabolites (organic acids, carbohydrates and amino acids) to the organ surface.3 Sugars such as sucrose, glucose and fructose are the most abundant carbon sources on the epidermal cells that have been investigated so far and presumably leach from the interior of the plant tissues. According to Lindow and Brandl4 about 0.2 to 10 µg of sugar, which is enough to support the growth of 107 to 108 bacteria per leaf, could be washed from undamaged, germ-free organs. Nevertheless, the intact phyllosphere must be viewed as an oligotrophic environment with respect to carbon- and nitrogen availability. The rate of nutrient leakage depends on several factors such as leaf age, intensity of dew and rain, or sun exposure. Moreover, several studies document that sugars are relatively abundant in only a few locations (“oases” of diluted organic nutrients); hence, most leaf microbes live in a variable, moist to dry habitat that is poor in dissolved sugars.4
Methanol Emission of Leaves
Land plants produce and emit a variety of volatile organic compounds (VOCs) such as isoprenoids (isoprene, monoterpene). In 1993 it was reported that tree and crop species emit substantial amounts of the VOC methanol (CH3OH).5 Based on a series of studies it was concluded that the tissues of all C3-plants release the C1-alcohol CH3OH, which is emitted primarily from stomates6,40 (Fig. 2). In addition, it is documented that young (growing) leaves emit more methanol than fully expanded organs.
Figure 2.
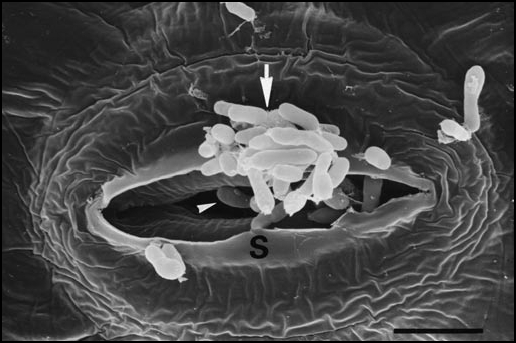
Methylobacteria as epiphytes and endophytes. Scanning electron micrograph of the surface of a leaf from a regenerated, germ-free sunflower (Helianthus annuus) plantlet cultured in vitro. The tissue was inoculated with a strain of Methylobacterium that was isolated from a free-living sunflower plant (M. sp.). A cluster of methylobacteria can be seen at the outer surface of a stomatum (S) (arrow). Single bacteria are visible in the pore of the stomatum and apparently invade the plant body (arrow head). Bar = 0.5 µm.
It has been shown that methanol formation in expanding plant cells is to large extend due to an enzymatic reaction restricted to the wall. Pectins, a heterogenous class of polymers, represent 20–35% of the dry mass of dicot walls.7 These components of the extracellular matrix are secreted via the Golgi apparatus in a highly methylesterified state. Within the wall space, they are de-esterified by a group of wall-associated enzymes, the pectin-methylesterases. In this reaction, the methoxy groups of the secreted pectins are converted into carboxyl groups, releasing the VOC methanol and protons.5 Experiments with growing pollen tubes (male gametophytes of angiosperms) support the concept that pectin-methylesterases are involved in the regulation of tip growth.6
All available evidence indicates that methanol is a waste product of the pectin metabolism in growing walls. Prokaryotic “waste managers” inhabit the outer surface of the cells (Fig. 1A and B); sometimes they actively move to and invade the pores of stomata and may establish endophytic bacterial populations. The extent of endophytic colonization of the sunflower plant is unknown (Fig. 2). It should be noted that it is likely that the methylobacteria are attracted by the emitted methanol toward the open stomata. However, direct proof for this assumption is currently lacking. The mobile methanol-consuming epiphytic microbes depicted in Figure 2 are described in the next section.
Free-Living Proteobacteria, Mitochondria and the Genus Methylobacterium
Plant-microbe interactions can be beneficial, neutral or detrimental to the photosynthetic (green) host organism8: from the “viewpoint” of the embryophyte, the prokaryotic microorganisms are either symbionts, commensals, or pathogens.9 Andrews and Harris10 presented a biogeographic model of the plant surface and a molecular phylogenetic classification of representative epiphytes. Based on the Ribosomal Database Project (small-subunit [16S] rRNA sequences) it became apparent that most epiphytic bacteria belong to the subdivision of Alpha-Proteobacteria (genera Rhizobium, Agrobacterium, Methylobacterium, etc.). These primarily free-living “purple bacteria” can form symbiotic relationships (rhizobia as nitrogen-fixing microbes in the root nodules of legumes11), act as plant pathogens (agrobacteria that induce crown-gall tumors12) or live as leaf-associated commensals/beneficial partners of the plant (methylobacteria as phytosymbionts3). It should be noted that some obligate intracellular parasites of animals and humans that can cause typhus (microbes of the genus Rickettsia) are also members of the Alpha-subclass of Proteobacteria.11 On the basis of rRNA sequence data it is documented that microbes of the genus Rickettsia belong to those extant bacteria that are most closely related to the mitochondria of eukaryotic cells.9
Ten years ago, M.A. Holland13,14 speculated that land plants (sporophytes) may have an ancient symbiotic relationship with cytokinin-producing epiphytes of the genus Methylobacterium. According to Green,15 Alpha-Proteobacteria of this genus can be defined as gram-negative, strictly aerobic, rod-shaped microbes that are able to grow on one-carbon compounds such as methanol or formaldehyde as well as on a variety of other organic substances. Since most “bacterial species” within this genus are red to pink due to the presence of carotenoids, methylobacteria are also referred to as pink-pigmented facultative methylotrophs (the validity of species concepts with respect to prokaryotic microbes is discussed in the ref. 16). The type strain is Pseudomonas mesophilica (= Methylobacterium mesophilicum). These microbes were isolated in May 1973 from the leaf surface of perennial ryegrass (Lolium perenne) and thereafter maintained in the laboratory.17 Methylobacteria are widely distributed in a variety of habitats, such as freshwater, dust and soils.4 On leaf surfaces, these methanol-consuming microbes often dominate the “bacterial flora” (microbiota)14: Quantitative studies led to the conclusion that typically more than 80% of the viable bacteria recovered from healthy leaves of representative embryophytes are members of the genus Methylobacterium.5,35
When Holland published his hypothesis, no direct evidence for the biosynthesis of cytokinins by plant cells was available13,14 (cytokinins are a class of phytohormones that regulate cell division activity in the meristems of the plant body). Accordingly, Holland proposed that cytokinins are produced by the extracellular microbial symbionts of embryophytes, and not by the plants themselves. Based on experimental results with soybeans (Glycine max), he concluded that the growth of the green plant (sporophyte) depends on cytokinins that are supplied by epiphytic microbes of the genus Methylobacterium. Hence, according to Holland, cytokinin-producing microbes are associated with all plants, and, as a corollary, in the absence of these epiphytes cell growth should be drastically reduced or impossible.13,14
Experiments with Sunflower Seedlings and “Living Fossil” Plants
In the year 2001 the author of this article discovered that healthy looking, rapidly growing seedlings of sunflower (Helianthus annuus) that were raised under non-sterile standard conditions7 are contaminated with epiphytic bacteria via the seed coat. The enclosed sunflower embryo is germ-free (sterile), but the outer surface of the pericarp is the habitat of a variety of microbes,18 which colonize the juvenile plant upon “infection” from without (seed coat). We have raised germ-free sunflower seeds under sterile conditions and compared the development of the resulting “gnotobiotic” seedlings with that of non-sterile samples. Germ-free sunflower seedlings grew significantly more rapid than the contaminated controls. Accordingly, it was concluded that epiphytes in the above-ground phytosphere are not required for the development of the sporophyte in this species.19
In a subsequent study we established an in vitro regeneration system using sunflower as model organism.20 Excised apical hypocotyl hook segments from etiolated seedlings were cultured on a shoot induction medium. Half of the hypocotyl segments were immersed in a Methylobacterium suspension (M. sp., an unidentified “species” isolated from a ligulate flower of a field-grown Helianthus annuus plant). The explants were cultured for four weeks. Shoots regenerated from the hypocotyl segments were cultured on a vitamin medium and thereafter transferred to a rooting solution. Regenerated H. annuus mini-plants started to flower after about 50 days of growth (stem length of the adult organisms: ca. 30 mm). Shoot induction and the percentage of rooted stems were significantly enhanced, but growth of the mini-sunflowers was not promoted by the methylobacteria. Since germ-free control plantlets developed normally it was concluded that the sunflower plant can produce sufficient amounts of cytokinins and other phytohormones, i.e., sporophyte development is not dependent on exogenously produced (bacterial) growth substances.20
In a subsequent study with germ-free seedlings of maize (Zea mays) and sunflower21 the same result was obtained: gnotobiotic juvenile plants of these species did not grow more rapidly in the presence of methylobacteria. In these experiments, the type-strain M. mesophilicum,17 a microbe that is a naturally occurring common resident of leaves, was used and compared with non-inoculated controls. These data disprove Holland's original hypothesis of 1997,13,14 at least with respect to the diploid sporophyte generation of Helianthus and Zea. Since the taxa used here are representative members of the angiosperms (di- and monocotyledonous land plants, respectively) it is likely that this general conclusion applies to all embryophytes. It should be noted that growth-promoting rhizobacteria (Bacillus subtilis etc.), i.e., microbes that colonize the root system of higher plants in nature, were not investigated. Their potential effect on organ development in sunflower and maize is still unexplored.22
In contrast to sunflower and maize, which are higher plants that are adapted to growth and reproduction in relatively dry regions of the biosphere, bryophytes (liverworts, mosses and hornworts) are ancient “living fossil” embryophytes23 that had never left their moist habitat and need droplets of water for fertilization.24,25 It has been known for some time that the growth of the haploid gametophytes in representative species of bryophytes is promoted by certain naturally occurring bacteria.26,27 However, a systematic analysis of this phenomenon was lacking. In order to fill this gap in our knowledge we analyzed the effect of methylobacteria on the growth of gametophytes of the moss Funaria hygrometrica and two distantly related species of liverworts, Marchantia polymorpha and Lunularia cruciata.
Protonemata (filamentous gametophytes) of the moss F. hygrometrica (Fig. 3A) were cultivated in vitro under germ-free conditions and their development recorded (changes in cell length and number of cells per filament). In the presence of three different “species” (strains) of methylobacteria (M. mesophilicum; M. sp. 1, isolated from phylloids of sporophytes of the moss F. hygrometrica, (Fig. 3B); and M. sp. 2, isolated from sunflower achenes) the rate of protonema growth was much higher than in the sterile control (Fig. 3C).28 A growth-promoting effect of similar magnitude was also recorded with uncontaminated gemmae (i.e., specialized propagules) isolated from mature gemma cups of the liverwort-species M. polymorpha and L. curciata. In the presence of methylobacteria (M. mesophilicum) the average surface area of developed gametophytes (expanded gemmae) was about three-fold larger than in the uncontaminated control.29
Figure 3.
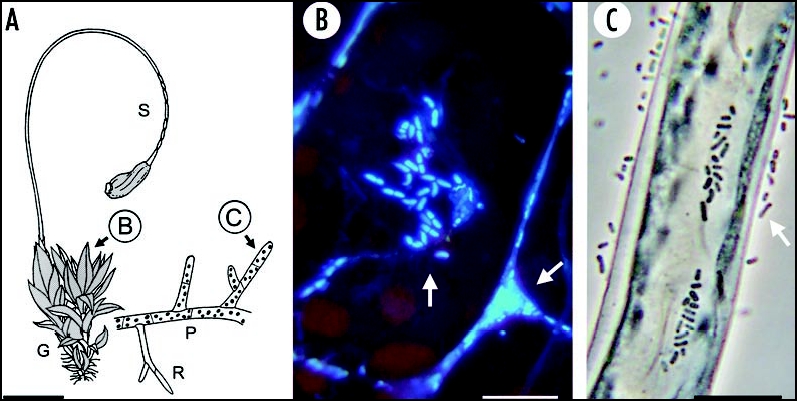
The moss Funaria hygrometrica (A): adult plantlet with the leafy gametophyte (G) and the stalk-like sporophyte (S); a filamentous protonema (P) with rhizoids (R) is also depicted. On the lamina of a leaflet (B) numerous epiphytic microbes are detectable by fluorescence microscopy (arrows). Representative protonema cell, 5 days after sowing of the spores, with methylobacteria that are free-living or attached to the surface of the wall (arrow) (C). Bars = 5 mm (A), 20 µm (B and C).
Production of Phytohormones by Epiphytic Methylobacteria
Using a variety of biochemical and cytological methods (immunoaffinity chromatography / radioimmunoassys; a specific bio-assay with axenic moss-protonemata) it has been shown that microbes of the genus Methylobacterium, maintained in liquid culture, produce cytokinin and secret it into the surrounding medium.29 The “classical” phytohormone auxin (indole-3-acetic acid) is also produced and secreted by different strains of methylobacteria.3,28 However, addition of the amino acid tryptophan, a precursor of auxin, was required for the stimulation and continuous biosynthesis of bacterial indole-3-acetic acid.30,31 In a detailed study it was shown that the rate of auxin production is strain-dependent and therefore a “species-specific” characteristic. Based on two independent methods (a colorimetric technique and thin layer chromatography) Hornschuh et al.3 have shown that the “domesticated” type strain M. mesophilicum17 that has been maintained in the laboratory since 1973, produces only about 1/3 the amount of auxin compared with two unidentified, plant-associated “wild-type” isolates of the genus Methylobacterium.
Taken together, these results document that non-pathogenic methylobacteria produce and release two important growth-promoting substances that are involved in the regulation of cell division activity32 and cell expansion33,34 in all land plants investigated so far. The implications of this finding are discussed in the last section of this article.
Conclusions
Botanists have long known that the outer surface of plants is large, whereas the internal cell-free areas of the body are relatively small (lumen of the xylem, intracellular spaces). In other words, “plants wear their guts on the outside”.10 In the Introduction it is described that germ-free (gnotobiotic) mice carry coevolved prokaryotic symbionts in their intestinal system, the so-called “gut microbes” of the mammals.1 The results summarized here are based on analogous studies carried out with gnotobiotic land plants that were inoculated with specific microbes. With respect to prokaryotes of the genus Methylobacterium and gametophytes of “living fossil” land plants (bryophytes) the following hypothetical plant-microbe-interaction is postulated (Fig. 4). Growing plant cells release metabolic waste products such as methanol5 and organic compounds (amino acids etc.) that are taken up and consumed by the bacteria.35,36,40 The epiphytes metabolize these materials and degrade them into ever simpler molecules (ammonium ions etc.) that are recycled by the plant cells. As a result of this continuous nutrition the methylobacteria produce and secrete cytokinin29 and auxin,3,31 which are signals to the plant indicating that the epiphytes are present. These exogenous phytohormones stimulate the growth and metabolism of the gametophyte (enhancement in the rate of cell division and cell expansion).
Figure 4.
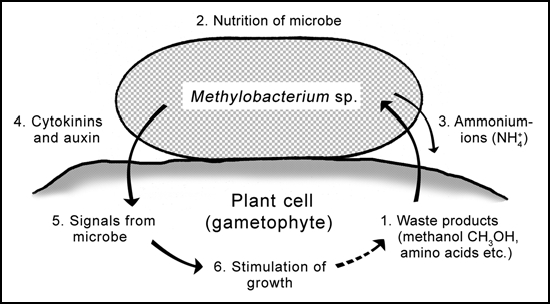
Naturally occurring epiphytic methylobacteria as coevolved symbionts of the gametophytes of ancient land plants (bryophytes). Growing plant cells produce waste products (methanol; leakage of amino acids etc.) (1) that are taken up and metabolized by the bacteria (2) which may release ammonium-ions (3). The extracellular prokaryotic “waste managers” produce and secrete cytokinins and auxin (4). These bacterial signals (5) may indicate to the plant that epiphytes are present and active. The exogenously produced phytohormones stimulate the growth of the gametophyte (6). This “cross-talk” between the eukaryotic host and the bacteria may contribute to the regulation of plant growth. Germ-free gametophytes (moss protonemata, liverwort thalli) develop at a much lower rate than “normal” (contaminated) individuals, which represent the situation found under natural conditions.
The first bacteria evolved about 3.5 billion years ago, whereas the earliest eukaryotic cells—products of a primary endosymbiosis—are much younger (less than 2 billion years).9,37 These facts show that eukaryotic (multicellular) macroorganisms such as plants and animals developed in an ancient world that was dominated by microbes, which also today represent the majority of living beings.2 When the first bryophyte-like plants colonized the moist areas around streams and lakes more than 400 million years ago (late Silurian), methylobacteria-like prokaryotes may already have been present.9 It is proposed here that the proto-methylobacteria associated with the earliest gametophytes of ancient bryophytes coevolved with their host organism.38 Hence, the microbe-plant-interaction depicted here (Fig. 4) must be interpreted as a very old symbiosis, i.e., methylobacteria are phytosymbionts. It should be noted that several years ago Y.A. Trotsenko et al.39 came to a similar conclusion. However, their data base was restricted to results obtained with sporophytes. The novel data summarized here show that growth, survival and reproductive success (i.e., fitness) of the gametophytes raised in the presence of methylobacteria is considerably higher than that of the germ-free plants. This “gnotobiotic minus control” is a laboratory artefact that does not occur under natural conditions, i.e., the inoculated samples represent the situation in the field (Fig. 3B and C).
It should be stressed that this hypothesis (Fig. 4) only applies to haploid gametophytes of “living fossil plants” such as mosses and liverworts (Fig. 3A). Our results do not support the concept that the growth of diploid sporophytes of higher plants such as maize and sunflower (Fig. 1A) is dependent on methylobacteria.13,14 This conclusion is in accordance with a recent study in which it was shown that seedling development in eight out of ten different species of higher plants is not significantly promoted by methylobacteria.40 Nevertheless, the bacterial epiphytes may have beneficial effects by preventing or reducing pathogenic microorganisms on the surface of the plant.41,42 It is concluded that the evolution of eukaryotes on Earth37,44 was to a large extent influenced by bacteria and that more work is required to further elucidate the role of microbes as a selective force in plant development and survival.43,44
Acknowledgements
I thank my co-workers R. Grotha, V. Koopmann, M. Hornschuh and S. Schauer for helpful comments on the manuscript and B. Teubert for technical assistance.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=4073
References
- 1.Bajzer M, Seeley RJ. Obesity and gut flora. Nature. 2006;444:1009–1010. doi: 10.1038/4441009a. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornschuh M, Grotha R, Kutschera U. Moss-associated methylobacteria as phytosymbionts: An experimental study. Naturwissenschaften. 2006;93:480–486. doi: 10.1007/s00114-006-0137-7. [DOI] [PubMed] [Google Scholar]
- 4.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fall R, Benson AA. Leaf methanol - The simplest natural product from plants. Trends Plant Sci. 1996;1:296–301. [Google Scholar]
- 6.Bosch M, Cheung AY, Hepler PK. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005;138:1334–1346. doi: 10.1104/pp.105.059865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutschera U. Cell expansion in plant development. Rev Brasil Fisiol Veg. 2000;12:65–95. [Google Scholar]
- 8.Steinert M, Hentschel J, Hacker J. Symbiosis and pathogenesis: Evolution of the microbe-host interaction. Naturwissenschaften. 2000;87:1–11. doi: 10.1007/s001140050001. [DOI] [PubMed] [Google Scholar]
- 9.Kutschera U, Niklas KJ. Endosymbiosis, cell evolution, and speciation. Theory Biosci. 2005;24:1–24. doi: 10.1016/j.thbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 11.Moulin L, Munive A, Dreyfus B, Boivin-Mason C. Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 12.Kutschera U, Bahrami M, Grotha R. Sucrose metabolism during Agrobacterium tumefaciens-induced tumor growth in sunflower hypocotyls. J Plant Physiol. 2000;157:1–6. [Google Scholar]
- 13.Holland MA. Occam's razor applied to hormonology: Are cytokinins produced by plants? Plant Physiol. 1997;115:865–868. doi: 10.1104/pp.115.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland MA. Methylobacterium and plants. Rec Res Dev Plant Physiol. 1997;1:207–213. [Google Scholar]
- 15.Green PN. The genus Methylobacterium. In: Balours A, Trüper HG, Dworkin M, Harder W, Schleifer KH, editors. The Prokaryotes. A Handbook on the Biology of Bacteria; Ecophysiology, Isolation, Identification, Applications. 2nd ed. Berlin Heidelberg New York: Springer; 1992. pp. 2342–2349. [Google Scholar]
- 16.Kutschera U. Species concepts: Leeches versus bacteria. Lauterbornia. 2004;52:171–175. [Google Scholar]
- 17.Austin B, Goodfellow M. Pseudomonas mesophilica, a new species of pink bacteria isolated from leaf surfaces. Int J Syst Bacteriol. 1979;29:373–378. [Google Scholar]
- 18.Kutschera U. Bacterial colonization of sunflower cotyledons during seed germination. J Appl Bot. 2002;76:96–98. [Google Scholar]
- 19.Kutschera U, Koopmann V, Grotha R. Plant development in the absence of epiphytic microorganisms. Naturwissenschaften. 2002;89:319–321. doi: 10.1007/s00114-002-0334-y. [DOI] [PubMed] [Google Scholar]
- 20.Koopmann V, Kutschera U. In-vitro regeneration of sunflower plants: Effects of a Methylobacterium strain on organ development. J Appl Bot. 2005;79:59–62. [Google Scholar]
- 21.Kutschera U, Koopmann V. Growth in liverworts of the Marchantiales is promoted by epiphytic methylobacteria. Naturwissenschaften. 2005;92:347–349. doi: 10.1007/s00114-005-0640-2. [DOI] [PubMed] [Google Scholar]
- 22.Compant S, Duffy B, Nowak J, Clément C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niklas KJ. The evolutionary biology of plants. Chicago and London: The University of Chicago Press; 1997. [Google Scholar]
- 24.Willis KJ, McElwain JC. The evolution of plants. Oxford and New York: Oxford University Press; 2002. [Google Scholar]
- 25.Scherp P, Grotha R, Kutschera U. Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Rep. 2001;20:143–149. doi: 10.1007/s002990000301. [DOI] [PubMed] [Google Scholar]
- 26.Basile DV, Slade LL, Corpe WA. An association between a bacterium and a liverwort, Scapania nemorosa. Bull Torrey Bot Club. 1969;96:711–714. [Google Scholar]
- 27.Spiess LD, Lippincott BB, Lippincott JA. Specificity of moss response to moss-accociated bacteria: Some influences of moss species, habitat, and locale. Bot Gaz. 1986;147:418–424. [Google Scholar]
- 28.Hornschuh M, Grotha R, Kutschera U. Epiphytic bacteria associated with the bryophyte Funaria hygrometrica: Effects of Methylobacterium strains on protonema development. Plant Biol. 2002;4:682–687. [Google Scholar]
- 29.Koenig RL, Morris RO, Polacco JC. tRNA is the source of low-level trans-Zeatin production in Methylobacterium spp. J Bacteriol. 2002;184:1832–1842. doi: 10.1128/JB.184.7.1832-1842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanova EG, Doronina NV, Trotsenko YA. Aerobic methylobacteria are capable of synthesizing auxins. Microbiology. 2001;70:392–397. [PubMed] [Google Scholar]
- 31.Omer ZS, Tombolini R, Broberg A, Gerhardson B. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Regul. 2004;43:93–96. [Google Scholar]
- 32.Haberer G, Kieber JJ. Cytokinins: New insights into a classic phytohormone. Plant Physiol. 2002;128:354–362. doi: 10.1104/pp.010773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutschera U. Auxin-induced cell elongation in grass coleoptiles: A phytohormone in action. Curr Top Plant Biol. 2003;4:27–46. [Google Scholar]
- 34.Kutschera U. Acid growth and plant development. Science. 2006;311:952–953. doi: 10.1126/science.311.5763.952b. [DOI] [PubMed] [Google Scholar]
- 35.Corpe WA, Rheem S. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol. 1986;62:243–250. [Google Scholar]
- 36.Holland MA, Polacco JC. PPFMs and other covert contaminants: Is there more to plant physiology than just plant? Annu Rev Plant Physiol Plant Mol Biol. 1994;45:197–209. [Google Scholar]
- 37.Kutschera U, Niklas KJ. The modern theory of biological evolution: An expanded synthesis. Naturwissenschaften. 2004;91:255–276. doi: 10.1007/s00114-004-0515-y. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JN. The coevolutionary process. Chicago: University of Chicago Press; 1994. [Google Scholar]
- 39.Trotsenko YA, Ivanova EG, Doronina NV. Aerobic methylotrophic bacteria as phytosymbionts. Microbiology. 2001;70:623–632. [Google Scholar]
- 40.Abanda-Nkpwatt D, Müsch M, Tschiersch J, Boettner M, Schwaab W. Molecular interaction between Methylobacterium extorquens and seedlings: Growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot. 2006;57:4025–4032. doi: 10.1093/jxb/erl173. [DOI] [PubMed] [Google Scholar]
- 41.Gourion B, Rossignol M, Vorholt JA. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci USA. 2006;103:13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulze-Lefert P, Robatzek S. Plant pathogens trick guard cells into opening gates. Cell. 2006;126:831–834. doi: 10.1016/j.cell.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Trewavas A. Plant intelligence. Naturwissenschaften. 2005;92:401–413. doi: 10.1007/s00114-005-0014-9. [DOI] [PubMed] [Google Scholar]
- 44.Futuyma DJ. Evolutionary biology. 3rd ed. Sunderland, Massachusetts: Sinauer Associates; 1998. [Google Scholar]


