Abstract
Arabidopsis hexokinase1 (HXK1) is a glucose sensor that regulates gene expression and plant growth and development. We have previously developed a high glucose (6%) assay based on the seedling developmental arrest to isolate and characterize the glucose-insensitive (gin) mutants. The analysis of gin2 as a null HXK1 mutant has revealed that the regulatory functions of HXK1 are distinct from its conventional role in glycolysis. In the Nov 3rd issue of Cell, we presented a new insight into the mechanism of HXK1-dependent glucose signaling. By combining proteomic and binary interaction screens, we discover two HXK1 unconventional partners (HUPs). HXK1 and HUPs form a core complex in the nucleus and directly regulate glucose-responsive gene expression and plant growth. As the 6% glucose assay is complicated by additional osmotic stress and nitrate signals, we have tested the gin2 and hup mutants using the 2% glucose assay. We believe that the new and more physiological glucose assay could help us better dissect the molecular mechanisms that link glucose regulation to diverse plant signaling pathways. Further functional analysis of gin mutants and the components in the novel nuclear HXK1 complex will provide more comprehensive mechanistic understanding of glucose sensing and signaling in plants.
Key Words: Hexokinase1 complex, HXK1 unconventional partners, glucose sensing and signaling, glucose-insensitive mutants
For any living organism, it is essential to rapidly and efficiently adjust its metabolism and physiology to the changes of nutrient availability and other environmental factors that perturb its metabolic balance. To do so, metabolites often serve as signaling molecules. Glucose is one of the ancient signaling metabolites that plays a fundamental regulatory role in physiology, metabolism, growth, development and gene expression from bacteria and yeasts to mammals and plants.1–4 In the autotrophic plants, glucose is central tothe regulation of many vital processes, such as the control of gene expression, cell proliferation and death, seedling, root, stem and inflorescence growth, leaf expansion and senescence, and seed development.4–7 Significantly, glucose downregulates photosynthetic gene expressions to suppress source activities, but often upregulates growth and respiration-related gene expression to enhance sink activities.6,7
To identify signaling components and elucidate intracellular pathways involved in glucose sensing and signaling, Arabidopsis glucose insensitive (gin) or glucose oversensitive (glo) mutants have been isolated based on glucose repression of cotyledon expansion, chlorophyll accumulation, and leaf and root development on high (6%) glucose and MS medium.8,9 The characterization of the Arabidopsis HXK1 mutants, gin2-1 and gin2-2 isolated from such screens, has provided compelling evidence for the role of HXK1 as a glucose sensor in regulating gene expression and plant growth.10 Many other gin and glo mutants are involved in plant hormone biosynthesis or signaling.4,8,9 For example, gin1, gin5 and gin6 are allelic to aba2, aba3 and abi4, respectively, involved in ABA biosynthesis and signaling,11,12 whereas gin4 is a new ctr1 allele in ethylene signaling.8 Little is known about the regulatory components that are directly connected with the HXK1-dependent glucose signaling mechanism.
We have long observed that Arabidopsis HXK1 can be expressed and detected in the nucleus of maize mesophyll protoplasts, in which we first discovered the global glucose repression of photosynthesis gene promoter activities.10,13 After confirming that a fraction of Arabidopsis HXK1 is located in the nucleus,14 we carried out nuclear-targeted proteomics analysis and the yeast two-hybrid mating assays to identify two HXK1 unconventional partners (HUPs): vacuolar H+-ATPase B1 (VHA-B1/HUP1) and a subunit of the 19S regulatory particle of proteasome (RPT5B/HUP2). Protein interaction analysis in vivo has further demonstrated that these two proteins form a nuclear-specific complex with HXK1. Importantly, the loss-of-function hup1 and hup2 mutants share a broad spectrum of glucose insensitive phenotypes with the gin2 mutant. Because the 6% glucose assay is confounded by the presence of osmotic stress and antagonistic effect of nitrate (40 mM in the MS medium),4,9,10 we have tested the possibility of developing a specific glucose-signaling assay using exogenous glucose closer to physiological level (2%) without nitrate. In the 2% glucose assay, hup1, hup2 and gin2 show similar insensitivity to glucose (Fig. 1), further verifying their genuine functions in glucose sensing and signaling.10
Figure 1.
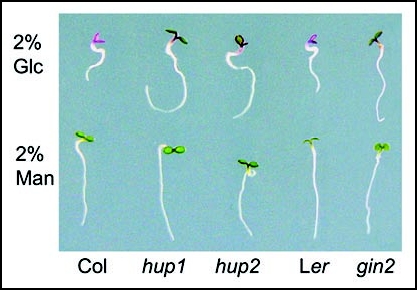
Glucose insensitivity of the hup1, hup2 and gin2 mutants. The three glucose-signaling mutants are insensitive to glucose-mediated developmental arrest observed in WT (Col and Ler). All plants display similar growth on the mannitol medium. The seedlings were grown on 2% glucose (Glc) or mannitol (Man) medium without MS for three days under constant light conditions (70 µmol/m2/s).
The gin2-like phenotypes of the hup1 and hup2 mutants are independent of HXK1 expression, HXK1 protein abundance, or glucose or fructose phosphorylation. Similar to HXK1, HUP1 and HUP2 are required for the glucose repression of photosynthetic genes (e.g., CAB and CAA). Chromatin immunoprecipitation assays have shown that the HXK1 protein complex is associated with the immediate 5′ upstream region of CAB2, where critical cis-elements have been identified.15 The association is enhanced during glucose repression and requires HUP1 and HUP2.14
Although the HXK1 complex is clearly enriched in the chromatin of early target genes and regulates transcription in the nucleus, its core components, HXK1, HUP1 and HUP2, do not contain DNA-binding activity. We have suggested that the recruitment of the nuclear HXK1 complex at glucose-responsive gene promoters may rely on their DNA-binding protein partners in the complex. Supporting this view, our characterization of the HXK1 complex has identified several putative transcription factors that interact directly with HUP1 and/or HUP2, but not with HXK1.14 Further analysis of these transcription factors and other complex components will provide a more comprehensive understanding of the function and mechanistic action of the nuclear HXK1 complex in the regulation of glucose responsive genes.
Abbreviations
- HXK1
hexokinase1
- HUP
HXK1 unconventional partner
- gin2
glucose-insensitive2
Footnotes
Previosly published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3894
References
- 1.Stulke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 2.Johnston M, Kim JH. Glucose as a hormone: Receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;3:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- 3.Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: Conserverd and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 5.Koch KE. Carbohydrate modulated gene expression in plant. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–515. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 6.Smeekens S. Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Gibson SI. Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Leon P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 9.Rook F, Bevan MW. Genetic approaches to understanding sugar-response pathways. J Exp Bot. 2003;54:495–501. doi: 10.1093/jxb/erg054. [DOI] [PubMed] [Google Scholar]
- 10.Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 11.Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arenas-Huertero F, Arroyo-Becerra A, Zhou L, Sheen J, León P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagisawa S, Yoo SD, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–525. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- 14.Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell BB, Andersson CR, Poole DS, Kay SA, Chory J. HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiol. 2003;133:1565–1577. doi: 10.1104/pp.103.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]


