Abstract
Phototropins (phot1 and phot2) are blue light-activated serine/threonine protein kinases that function to mediate a variety of adaptive processes that serve to optimize the photosynthetic efficiency of plants and thereby promote their growth. Light sensing by the phototropins is mediated by a repeated motif located within the N-terminal region of the protein designated the LOV domain. Although phototropins possess two LOV photosensors (LOV1 and LOV2), recent biophysical and structure-function analyses clearly indicate that the LOV2 domain plays a predominant role in regulating phototropin kinase activity owing to specific protein changes that occur in response to LOV2 photoexcitation. In particular, the central β-sheet scaffold plays a role in propagating the photochemical signal generated from within LOV2 to protein changes at the surface that are necessary for kinase activation.
Key words: phototropin, LOV domain, FMN, cysteinyl adduct, amphipathic helix, receptor autophosphoryation
Light-Induced Activation of Phot1
The primary amino acid structures of plant phototropins (phot1 and phot2) can be separated into two distinct regions: a N-terminal photosensory region linked to a C-terminal effector domain that includes a classic serine/threonine kinase motif.1–3 The N-terminal photosensory region comprises two LOV domains each of which binds the blue light absorbing cofactor flavin mononucleotide (FMN) as chromophore.4,5 LOV domains exhibit significant homology to motifs found in a diverse range of eukaryotic and prokaryotic proteins involved in sensing Light, Oxygen or Voltage, hence the acronym LOV.6
Bacterially expressed LOV domains are photochemically reactive as monitored by absorbance spectroscopy.7 In the dark, LOV domains bind FMN non-covalently forming a spectral species, LOV447, which absorbs maximally near 447 nm.7,8 Irradiation of the domain induces the formation of a covalent bond between the C(4a) carbon of the FMN and the sulfur atom of a nearby, conserved cysteine residue within the domain. Cysteinyl adduct formation occurs within microseconds of illumination and produces a spectral species, LOV390, that absorbs maximally near 390 nm.8
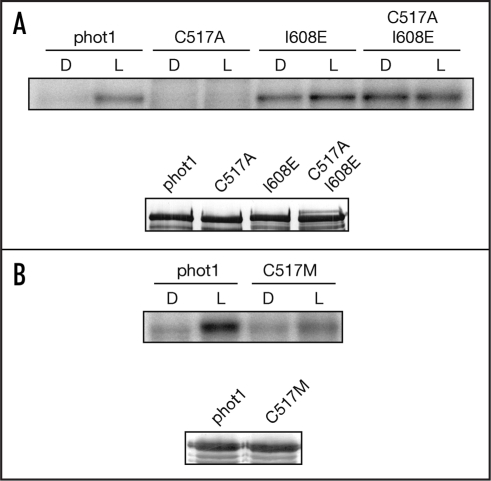
Mutation of the afore-mentioned active-site cysteine to either alanine or serine results in a loss of photochemical reactivity of the LOV domain.7,8 Moreover, this mutation has been used successfully to probe the roles of LOV1 and LOV2 in regulating phototropin activity and function.9–12 As reported previously (ref. 13), the Cys → Ala mutation within LOV2 of Arabidopsis thaliana phot1 (C517A) results in a loss of light-induced receptor autophosphorylation when expressed in insect cells (Fig. 1A), demonstrating that LOV2 is the principle photosensor regulating phot1 kinase activity. A similar mode of action has been reported for the LOV2 domain of Arabidopsis phot2.12,14,15 The LOV1 domain, by contrast, is proposed to mediate receptor dimerization16 and/or modulate the photoreactivity of LOV2.14
Figure 1.
Effect of point mutation analysis on the autophosphorylation activity of Arabidopsis phot1. (A) Autoradiograph showing light-dependent autophosphorylation activity of single C517A and I608E mutations and the combined double mutation in soluble protein extracts prepared from insect cells. All manipulations were completed under dim red light. Samples were given a mock irradiation (D) or irradiated with white light (L) at a total fluence of 30,000 µmol m-2 prior to the addition of radiolabelled ATP as described previously (refs. 13 and 20). Western blot analysis of phot1 protein levels using anti-phot1 antibody is shown below. (B) Autoradiograph showing the effect of the single C517M point mutation on phot1 autophosphorylation activity in soluble protein extracts prepared from insect cells. Western blot analysis of phot1 protein levels using anti-phot1 antibody is shown below.
Substitution of the conserved active-site cysteine with methionine has been shown to result in the formation of a unique photoproduct species absorbing maximally in the red region of the spectrum.17 This species is stable both under aerobic and denaturing conditions and consists of a covalent adduct between the introduced methionine and the N5 nitrogen of the FMN.17 We have found that an equivalent mutation within the LOV2 domain of Arabidopsis phot1 (C517M) does not completely abolish light-induced receptor autophosphorylation when expressed in insect cells (Fig. 1B). These findings imply that formation of a N5 methionyl adduct within the LOV2 domain still has the capacity to function as a signalling state, albeit at reduced capacity.
Kinase Regulation by LOV2
The current view of phototropin receptor activation is that LOV2 functions as a repressor of the C-terminal kinase domain in the dark and that this mode of repression is alleviated upon photoexcitation, resulting in receptor autophosphorylation.1,2 The sites of phototropin autophosphorylation have been mapped upstream of the LOV2 domain.18 However, there is still no information as to the functional consequences following receptor autophosphorylation. A truncated version of phot2 comprising only the LOV2 domain and the C-terminal kinase domain can restore functional activity in a phot2 mutant of Adiantum capillus-veneris.10 Thus, autophosphorylation may not be required for receptor signaling but may be involved in some other function, such as receptor desensitization.
The predominant role of LOV2 in regulating phototropin activity appears to arise from its position within the phototropin molecule. Photoexcitation of an extended LOV2 fragment leads to displacement of an α-helix from the surface of the LOV2-core.19 This α-helix, designated Jα, is amphipathic and situated downstream of LOV2. Substitution of non-polar amino acids located within the hydrophobic docking interface with polar residues results in constitutive disordering of the Jα-helix from the LOV2-core.20 Moreover, incorporation of these mutations into full-length Arabidopsis phot1 increases dark levels of autophosphorylation relative to wild-type controls.20 One mutation in particular, I608E, shows comparable levels of kinase activity in the dark and the light (Fig. 1A). In addition, the I608E mutation can restore phot1 kinase activity in the C517A mutant background (Fig. 1A), further demonstrating that unfolding of the Jα-helix results in activation of the C-terminal kinase domain. Further analysis is now required to assess the functional consequences of the I608E mutation on phototropin activity in planta.
LOV2 Signal Transmission
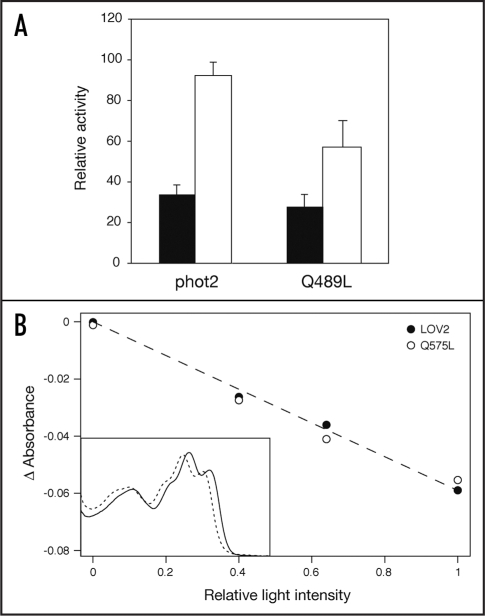
Biophysical studies have implicated a role for the central β-sheet scaffold in propagating the signal generated within the FMN-binding pocket to protein changes at the LOV2 surface, which are necessary for activation of the C-terminal kinase domain.21,22 Specifically, the side chain of a conserved glutamine residue within LOV2 (Gln575 in Arabidopsis phot1) which forms hydrogen bonds with the FMN chromophore flips by 180° upon cysteinyl adduct formation11,23,24 causing protein changes in the central β-sheet scaffold that forms contacts with the Jα-helix.19,20 We have recently shown that mutation of the conserved glutamine to leucine (Q575L) attenuates light-induced autophosphorylation of Arabidopsis phot1 expressed in insect cells,13 suggesting that this residue plays a role in transmitting the signal generated upon light-driven cysteinyl adduct formation from within LOV2 to protein changes at the LOV2 surface. An equivalent mutation when introduced into Arabidopsis phot2 (Q489L) also attenuates light-induced autophosphorylation (Fig. 2A) implying that the role of the conserved glutamine in LOV2 signal transmission is conserved between phototropins.
Figure 2.
Analysis of Gln→Leu mutations in full-length Arabidopsis phot2 and the LOV2 domain of Arabidopsis phot1. (A) Effect of the Q489L mutation on phot2 autophosphorylation activity in soluble protein extracts prepared from insect cells. Sample preparation and experimental procedures were performed as described in Figure 1. Kinase activity was quantified by phosphorimaging and expressed as a percentage of maximal autophosphorylation (errors bars indicate standard error). (B) Relative bleaching measurements for FMN-cysteinyl adduct formation using bacterially expressed phot1 LOV2 and the Q575L mutant. Light-induced absorbance changes were monitored at 448 nm (LOV2) or 440 nm (Q575L) over a range of light intensities as described previously (ref. 15). The fitted line gives a measure of relative bleaching for both proteins. Relative bleaching values for each protein are shown and represent the average of two independent protein preparations. Inset shows absorption spectra of phot1 LOV2 (solid line) and the Q575L mutant (dashed line) before bleaching.
It should be noted that incorporation of the Q575L mutation into the isolated LOV2 domain of Arabidopsis phot1 alters its spectral and photochemical properties relative to wild type, presumably due to a loss of hydrogen bonding with the FMN chromophore.13,21 The effects observed for the Q575L mutation on phototropin kinase activity may therefore result from a reduction in the photocycle quantum yield of LOV2. However, the rate of bleaching for bacterially expressed LOV2 harboring the Gln → Leu mutation, in response to pulses of light given at different intensities, is identical to that measured for the wild-type domain (Fig. 2B). The degree of bleaching for each domain is proportional to the light intensity, indicating that the light intensity used is not saturating.15 Hence, these measurements indicate that the quantum yield for adduct formation is the same for both wild-type LOV2 and the Q575L mutant. This conclusion is consistent with a structural relay mechanism whereby Gln575 is required to propagate conformational changes at the β-sheet surface upon LOV2 photoexcitation which in turn leads to Jα-helix displacement and subsequent activation of the C-terminal kinase domain.
A similar glutamine flipping signal transmission mechanism has recently been reported for the fungal LOV-containing photoreceptor, VIVID from Neurospora crassa.25 LOV-domain β-sheet rearrangement also appears to be required for interdomain interactions in the bacterial photosensor YtvA from Bacillus subtilis.26 Further studies will determine whether a signal transmission mechanism involving the central β-sheet is common to other LOV-sensor proteins. Interestingly, molecular dynamic simulations indicate that the consequences of LOV1 photoexcitation differ to that of LOV2 and involve stabilizing a conserved surface salt bridge.27 Mutational analysis of the salt bridge associated with LOV1 may provide additional insights as to the function of this domain, which to date remains poorly understood.
Abbreviations
- phot
phototropin
- LOV
Light, Oxygen or Voltage
- FMN
flavin mononucleotide
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/4848
References
- 1.Matsuoka D, Iwata T, Zikihara K, Kandori H, Tokutomi S. Primary processes during the light-signal transduction of phototropin. Photochem Photobiol. 2007;83:122–130. doi: 10.1562/2006-03-29-RA-861. [DOI] [PubMed] [Google Scholar]
- 2.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 3.Briggs WR. The LOV domain: A chromophore module servicing multiple photoreceptors. J Biomed Sci. 2007;14:499–504. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 4.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc Nat Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 6.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 7.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 8.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 9.Kanegae T, Hayashida E, Kuramoto C, Wada M. A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proc Natl Acad Sci USA. 2006;103:17997–18001. doi: 10.1073/pnas.0603569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M. Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol. 2004;45:416–426. doi: 10.1093/pcp/pch045. [DOI] [PubMed] [Google Scholar]
- 11.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho HY, Tseng TS, Kaiserli E, Sullivan S, Christie JM, Briggs WR. Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 2007;143:517–529. doi: 10.1104/pp.106.089839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of Phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka D, Tokutomi S. Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc Nat Acad Sci USA. 2005;102:13337–13342. doi: 10.1073/pnas.0506402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 16.Salomon M, Lempert U, Rudiger W. Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Letts. 2004;572:8–10. doi: 10.1016/j.febslet.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 17.Kottke T, Dick B, Fedorov R, Schlichting I, Deutzmann R, Hegemann P. Irreversible photoreduction of flavin in a mutated Phot-LOV1 domain. Biochemistry. 2003;42:9854–9862. doi: 10.1021/bi034863r. [DOI] [PubMed] [Google Scholar]
- 18.Salomon M, Knieb E, von Zeppelin T, Rudiger W. Mapping of low- and high-fluence autophosphorylation sites in phototropin 1. Biochemistry. 2003;42:4217–4225. doi: 10.1021/bi027324f. [DOI] [PubMed] [Google Scholar]
- 19.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 20.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J Am Chem Soc. 2004;126:3390–3391. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 21.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 22.Iwata T, Nozaki D, Tokutomi S, Kandori H. Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry. 2005;44:7427–7434. doi: 10.1021/bi047281y. [DOI] [PubMed] [Google Scholar]
- 23.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 24.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc Nat Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losi A, Ghiraldelli E, Jansen S, Gartner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem Photobiol. 2005;81:1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
- 27.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91:3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]