Abstract
Flowering is one of the most important steps in a plant life cycle. Plants utilize light as an informational source to determine the timing of flowering. In Arabidopsis, phytochrome A (phyA), phyB and cryptochrome2 (cry2) are major photoreceptors that regulate flowering. These photoreceptors perceive light stimuli by leaves for the regulation of flowering. A leaf is an organ consisting of different tissues such as epidermis, mesophyll and vascular bundles. In the present study, we examined in which tissue the light signals are perceived and how those signals are integrated within a leaf to regulate flowering. For this purpose, we established transgenic Arabidopsis lines that expressed a phyB-green fluorescent protein (GFP) fusion protein or a cry2-GFP fusion protein in organ/tissue-specific manners. Consequently, phyB was shown to perceive light stimuli in mesophyll. By contrast, cry2 functioned only in vascular bundles. We further confirmed that both phyB-GFP and cry2-GFP regulated flowering by altering the expression of a key flowering gene, FT, in vascular bundles. In summary, perception sites for different spectra of light are spatially separated within a leaf and the signals are integrated through the inter-tissue communication.
Key words: photoreceptor, light, flowering, phytochrome, cryptochrome, inter-tissue signal
The timing of flowering is strictly regulated by environmental conditions such as light. Two aspects of light, spectral nature and photoperiod, dramatically affect flowering. In Arabidopsis, phyB and phyA/cry2 are the major photoreceptors mediating these responses. Although photoreceptors are expressed in almost all organs,1 partial irradiation and grafting analyses have demonstrated that plants perceive light signals only in leaves.2–4 However, roles for different tissues in a leaf remained unknown due to a lack of a proper method. To answer the question, we established Arabidopsis transgenic lines that expressed phyB-GFP or cry2-GFP on the respective mutant backgrounds. The resultant transgenic lines were examined for their flowering phenotype. Consequently, we found that phyB-GFP in mesophyll but not in other tissues regulated flowering.5 By contrast, cry2-GFP functioned only in vascular bundles.6
A strong genetic interaction between phyB and cry2 in the regulation of flowering is known.7,8 Cry2 regulates the flowering by suppressing the inhibitory effect of phyB on flowering. Hence, cry2 function is observed only in the presence of phyB. Conversely, the effect of phyB is exaggerated in the cry2 mutant, because phyB is not counteracted by cry2 in its absence. Here, we tested how phyB and cry2 in different tissues regulated flowering in the absence of the other photoreceptor. For this purpose, we took a physiological approach. Phenotype of the phyB-GFP lines was examined under monochromatic red light, in which phyB but not cyr2 is activated. As expected, phyB-GFP in mesophyll but not in vascular bundles strongly affected the flowering in this condition (Fig. 1A). We also tested the cry2-GFP function when phyB was not activated. Namely, plants were placed under blue light supplemented with strong far-red light. As expected, cry2-GFP failed to affect the flowering even under this condition regardless of where it was expressed (Fig. 1B).
Figure 1.
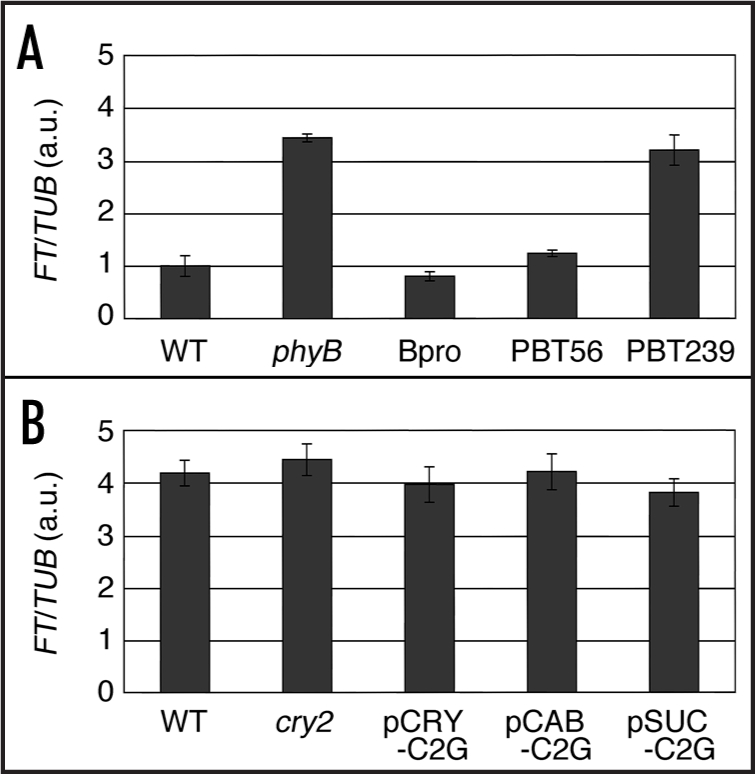
FT expression under phyB or cry2 inactive conditions. Total RNA was extracted from the seedlings grown under long-day condition for 10 days and subjected to qRT-PCR for FT expression analysis. Data were normalized to the level of FT mRNA in (A) of the wild type, which was set to 1 arbitrary unit (a.u.). Mean ± SE (n = 4). WT, wild type. (A) Long-day red light, (16L 8D; 10 µmol m-2 s-1). WT, wild type; phyB, phyB mutant; Bpro, PHYB promoter-PHYB-GFP; PBT56, phyB-GFP in mesophyll; PBT239, phyB-GFP in vascular bundles.5 (B) Long-day blue and far-red light (16L 8D; blue light, 3 µmol m-2 s-1; far-red light, 10 µmol m-2 s-1). WT, wild type; cry2, cry2 mutant; pCRY-C2G, CRY2 promoter-CRY2-GFP; pCAB-C2G, CAB3 promoter-CRY2-GFP; pSUC-C2G, SUC2 promoter-CRY2-GFP.6
Photoreceptors regulate flowering by altering the expression of a key flowering regulator, FT.9,10 Interestingly, the FT gene is expressed specifically in vascular bundles.11 Indeed, mesophyll phyB-GFP controlled the expression of FT in vascular bundles. Hence, there must be a mechanism by which the light signal is transduced from mesophyll to vascular bundles to regulate the FT expression in vascular bundles. It should be noted here that FT is not the sole factor involved in the light regulation of flowering. Factors such as CO, SPA, COP1 and PFT1 are known to link the photoreceptors and FT.12–14 These factors most likely function in leaves. However, their function sites at the tissue level remain totally unknown except for CO. The biological clock is another class of machinery that is tightly related to the light signal transduction pathway.15 Unfortunately, function sites of the clock components for the regulation of flowering remain unclear. The future work should reveal those sites. Such analyses should finally provide a complete picture illustrating a network of the inter-tissue signaling for the regulation of flowering.
The present work urges us to indentify the molecule that mediates the inter-tissue signaling between mesophyll and vascular bundles. Potential candidates include phytohormones, microRNA16 and peptides.17 Among phytohormones, gibberellin promotes flowering.18 However, gibberellin is probably not the answer because gibberellin does not alter the FT expression directly. Except gibberellin, no exogenously added phythromone dramatically affects flowering in Arabidopsis. It is known that microRNA such as miR172, miR159 and miR156 are involved in the regulation of flowering time.19 However, those microRNA's neither regulate the FT expression nor are regulated by light. Since most of microRNA's has not been intensively studied yet, it remains possible that one of them may mediate the above inter-tissue signal. Another potential candidate is a peptide. Although not much is known about peptide hormones in plants yet, peptides such as PSK,20 xylogen21 and CLE22 have been shown to regulate cell growth and differentiation. Although none of peptides is known to regulate flowering in plants at present, a future work may reveal a novel peptide that mediates the inter-tissue signals for flowering.
Acknowledgement
This work was partially supported by a Grant-in-Aid for Scientific Research (B) 17370018 (to A.N.), a Grand-in-Aid for Research on Priority Areas 17084002 (to A.N.), and a Grand-in-Aid for Japan Society for the Promotion of Science Fellows 02709 (to M.E.).
Abbreviations
- PHY
phytochrome
- CRY
cryptochrome
- GFP
green fluorescent protein
- FT
flowering locus T
and
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/4863
References
- 1.Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knott JE. Effect of a localized photoperiod on spinach. Proc Am Soc Hortic Sci. 1934;31:152–154. [Google Scholar]
- 3.Chailakhyan MK. On the hormonal theory of plant development. Dokl Akad Nauk SSSR. 1936;12:443–447. [Google Scholar]
- 4.Zeevaart JAD. Physiology of flower formation. Annu Rev Plant Physiol. 1976;27:321–348. [Google Scholar]
- 5.Endo M, Nakamura T, Araki T, Mochizuki N, Akira N. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell. 2005;17:1941–1952. doi: 10.1105/tpc.105.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo M, Mochizuki N, Suzuki T, Akira N. CRYPTOCHME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell. 2007;19:84–93. doi: 10.1105/tpc.106.048157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;27:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 8.Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- 9.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 11.Takada S, Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 14.Cerdan PD, Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- 15.Hayama R, Coupland G. Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol. 2003;6:13–19. doi: 10.1016/s1369-5266(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 16.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7:512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher KL, Benfey PN. Not just another hole in the wall: Understanding intercellular protein trafficking. Genes Dev. 2005;19:189–195. doi: 10.1101/gad.1271005. [DOI] [PubMed] [Google Scholar]
- 18.Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- 19.Quesada V, Dean C, Simpson GG. Regulated RNA processing in the control of Arabidopsis flowering. Int J Dev Biol. 2005;49:773–780. doi: 10.1387/ijdb.051995vq. [DOI] [PubMed] [Google Scholar]
- 20.Matsubayashi Y, Yang H, Sakagami Y. Peptide signals and their receptors in higher plants. Trends Plant Sci. 2001;6:573–577. doi: 10.1016/s1360-1385(01)02148-3. [DOI] [PubMed] [Google Scholar]
- 21.Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]


