Abstract
Plants have developed robust signaling mechanisms to cope with environmental stresses. In a recent article in the Plant Journal,1 we reported on a salt stress signaling pathway in Arabidopsis, which is mechanically similar to endoplasmic reticulum (ER) stress responses described in mammalian systems. We found that a type II membrane-bound transcription factor (AtbZIP17) is a salt stress signal transducer and a subtilisin-like serine protease (AtS1P) is a key component of the protein processing machinery in the signaling pathway. AtbZIP17 is normally located in the ER, but in response to salt stress, the membrane-bound transcription factor is proteolytically processed in an AtS1P-dependent step, and the cytosolic component of AtbZIP17 is translocated to the nucleus. We have demonstrated in an in vitro assay that AtS1P directly cleaves AtbZIP17, an initial step in processing. Our study also identified several downstream target genes in the salt signaling pathway. In this addendum, we propose a testable model to describe the action of AtS1P and AtbZIP17 in salt stress signaling and we also present an outlook for future work to test this model.
Key words: Arabidopsis, salt stress, transcription factor, proteolytic processing, nuclear relocation
Some of the most fascinating signaling systems in eukaryotes are those that respond to ER stress. ER stress occurs when normal protein folding or secretory processes are inhibited, and unfolded or misfolded proteins accumulate in the ER.2 What makes these systems so interesting is their cell biology. The transduction of ER stress signals involves the translocation of signaling components from one organellar compartment to another and activation by proteolytic processing during the translocation process. We have described an ER stress response in Arabidopsis that responds to salt stress.2 The salt stress response in Arabidopsis has elements in common with other well described ER stresses, such as the unfolded protein response (UPR) studied in yeast and mammalian cells and cholesterol homeostasis described in humans.
The cell biology of ER stress signaling first came to light through the elegant studies of Brown and Goldstein on the control of cholesterol biosynthesis in mammalian systems.3 The operation of the pathway revolves around the activation and organelle translocation of sterol regulatory element binding proteins (SREBPs). SREBPs are bHLH transcription factors resident in the ER where they interact with SREBP cleavage-activating protein (SCAP), a membrane protein having a sterol-sensing domain and binding to a protein encoded by the insulin-induced gene (Insig).4 Under sterol deprivation conditions, SCAP is released from its interaction with Insig and escorts SREBP to the Golgi apparatus where SREBP is activated by the protein processing machinery resident in that organelle.5 The SREBP transcriptional activation domain is translocated to the nucleus where it binds to and activates the expression of genes in cholesterol biosynthesis.
UPR signaling is a similar process—triggered by the accumulation of unfolded or misfolded proteins in the ER.6 UPR can be activated by stress agents that prevent proper protein folding, but is thought to be a general stress response since protein folding is exquisitely sensitive to environmental stress. UPR in mammalian systems is mediated by several ER membrane sensor/transducers. The one most relevant to the system described in our paper is activating transcription factor 6 (ATF6). ATF6, like the SREBPs, is a trans-membrane transcription factor resident in the ER with a bZIP domain on its N-terminus or cytosolic facing domain.7 The C-terminus of the protein is lumen facing and is thought to be tethered in the ER by its interaction with Binding Protein (BiP).8 When BiP is recruited for interaction with misfolded proteins in the ER during UPR, ATF6 is released to the Golgi apparatus where it encounters the proteolytic processing machinery.
We stumbled across a similar signaling system in plants, through functional studies of subtilisin-like serine proteases in Arabidopsis. There are 56 genes encoding these proteases in Arabidopsis and one of them, AtS1P, encodes a protein with remarkable similarity to S1P, a key component of the proteolytic machinery that processes SREBP and ATF6 in mammals.9 At the time, we did not know what were substrates for AtS1P and whether AtS1P was involved in stress signaling. We found that at least two T-DNA knockout lines of AtS1P were salt sensitive and suspected from this that AtS1P was involved in salt stress responses. To identify components of a signaling pathway upon which AtS1P might be acting, we looked for genes in the Arabidopsis genome encoding bZIP transcription factors with structural similarities to ATF6—a protein with an N-terminal bZIP domain, a -trans-membrane domain and a canonical S1P site in the C-terminal domain.7 We found three genes encoding AtbZIP17, -28 and 49 and from these we created epitope-tagged versions of AtbZIP17 and -28. We observed that AtbZIP17 was processed in response to salt stress and that the processing was AtS1P dependent. We found that epitope-tagged versions of AtbZIP17 were localized in the ER and relocated to the nucleus in response to salt stress.
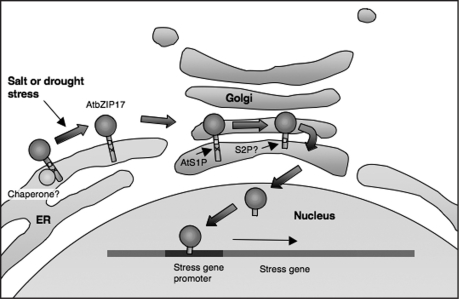
Further work is required to fully understand how this system works under salt stress. We would like to propose a simple testable model that could account for the results reported in this paper (Fig. 1). We assume, but have not yet been able to show definitely that AtbZIP17 follows the same organelle translocation route as the SREBPs and ATF6. We know that AtS1P is localized in the Golgi, but so far we have not been able to catch AtbZIP17 en route through that organelle. It may be that the residence time in the Golgi is brief, so we plan to examine AtbZIP17 translocation in an AtS1P mutant in which processing is blocked. We also know from the size of the final product that AtbZIP17 is further processed by other protease(s). We are looking to see whether AtbZIP17 is processed by a site-2 protease (S2P), a Zn2+ metalloprotease, as are SREBP and ATF6.9
Figure 1.
A model is shown for the activation of AtbZIP17 in response to salt stress. AtbZIP17 is a membrane-associated bZIP transcription factor normally located in the ER. In response to salt stress, AtbZIP17 is activated and presumably migrates to the cis-medial Golgi. AtS1P, a subtilisin-like serine protease, resident in the Golgi, initiates the proteolytic processing by clipping the C-terminal tail of AtbZIP17. Processing continues by other unidentified proteases, releasing the cytosolic-facing component of AtbZIP17, which relocates to the nucleus activating salt stress response genes.
We don't know what is the salt stress sensor. Is the salt stress response just a glorified UPR responding to a different class of misfolded proteins under salt stress? We know that salt stress is not a standard UPR, because stress agents such as tunicamycin activate another bZIP transcription factor and not AtbZIP17 (Liu et al., manuscript submitted), and typical UPR downstream target genes are not upregulated by salt stress in our experiments. Is there a factor, such as SCAP or BiP that interacts with AtbZIP17 and acts as a salt stress sensor? Does AtbZIP17 sense these conditions on its own? Perhaps the system does not sense salt stress directly at all but senses the action or product of another pathway that does.
There are many salt stress signaling pathways and salt stress responses that have already been described in plants, such as SOS pathway, ABA-dependent responses and ABA-independent responses (for example, see ref. 10). What are the relationships of these responses to the one we have found? A hint of what is going on may be revealed by the genes activated by AtbZIP17. We have identified genes dependent on AtS1P in response to salt stress and among those we have verified that they are likewise dependent on AtbZIP17. We do not yet know whether these genes are direct targets of AtbZIP17, but the list does include a transcription factor ATHB-7 that has been shown by others to be salt stress activated.11
We could be further enlightened about AtbZIP17 function by expressing truncated forms of the gene in transgenic plants. Truncated forms expressing only the cytosolic component of AtbZIP17 should bypass the need for proteolytic processing. Such plants should reveal the consequence of constitutive activation of this stress response system.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/4889
References
- 1.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to ER stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Kaufman RJ. A trip to the ER: Coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 4.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 7.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 9.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 10.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 11.Soderman E, Mattsson J, Engstrom P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996;10:375–381. doi: 10.1046/j.1365-313x.1996.10020375.x. [DOI] [PubMed] [Google Scholar]