Abstract
Appropriate embryonic patterning is amongst the most fundamental processes in plant development, necessary for the correct specification of root and shoot apical meristems which generate all post-germination organs of a plant. Many mutations have been characterized which disrupt embryonic pattern formation and many recent studies have focussed on the role of auxin in establishing apical-basal polarity. Our recent work has demonstrated the role of two redundant AP2 transcription factors, DORNROESCHEN (DRN) and DORNROESCHEN-LIKE (DRNL) in the control of embryo patterning, upstream of auxin perception and/or response and that DRN in turn, is regulated by auxin. We also suggest both genes are involved in the change from radial to bilateral symmetry in the globular embryo and are responsible for positional information of meristem-specific genes such as STM. The promiscuous interaction of DRN and DRNL proteins with the redundant family of class III HD-ZIP partners may represent a way by which embryonic cell specification can be controlled by combinations of transcription factor complexes, together with auxin.
Key words: embryo, pattern formation, auxin, cotyledon, Arabidopsis
Embryonic Patterning is Under Redundant Control
The elaboration of an embryo from a fertilized zygote into a fully differentiated seed is achieved by processes of cell division and cell differentiation. In Arabidopsis, patterning events involving the initiation of apical-basal and radial axes and stem cell production. Differentiation ensues from a highly characteristic and reproducible pattern of cell divisions throughout embryogenesis, which has enabled the identification of mutants with aberrant patterns. Many mutants have been reported which mainly either affect the apical domain, giving rise to altered cotyledon morphogenesis, or the basal domain, resulting in root defects. The low penetrance of embryo phenotypes resulting from single mutations within many gene families demonstrates that redundancy is an important feature of the control of embryo patterning. Dynamic changes in auxin concentration have been implicated in patterning processes including the establishment of apical-basal polarity and bilateral symmetry.1,2 Correspondingly, patterning phenotypes arise from mutations in genes involved in auxin signalling or transport such as PINOID,3 or within the PIN family.1,2 Additionally, mutations in the auxin response factor MONOPTEROS (MP) or its IAA inhibitor BODENLOS (BDL),4,5 the CUC gene family,6–8 or higher order mutations within the class III HD-ZIP family of transcription factors,9 result in disrupted embryonic patterning and effects on polar auxin transport.10
DRN/DRNL Redundantly Affect Apical/Basal Polarity Via Auxin
We recently showed that the DORNROESCHEN (DRN) and paralogous DORNROESCHEN-LIKE (DRNL) genes redundantly specify correct cotyledon development and cell divisions, particularly in the hypophysis region, implicating the AP2 class of transcription factors in embryonic patterning.11 Both genes alter apical/basal polarity by causing cell division defects in the basal embryo domain or cotyledon defects in the apical domain. Since expression of both genes is confined to the apical region, non-cell autonomous functions may be inferred, in a similar way to MP/BDL function,12 with auxin being one putative signal. Several lines of evidence currently implicate a role for DRN in auxin perception/response. Firstly, the expression of two markers for auxin concentration/response, DR5::GFP and PIN1::PIN1-GFP is altered in a drn mutant background, suggesting DRN functions upstream of auxin response/polar transport. Secondly, mono- or polycotyledonous drn and drnl mutants have altered leaf phyllotaxis, known to be dependent on local concentrations of auxin.13 Additionally, DRN upstream and downstream regulatory sequences contain several canonical auxin response elements (AREs) specifically targeted by auxin response factors (ARFs),14 (Fig. 1A), which we have shown to be functional by mutational analysis and responsible for auxin-driven DRN expression in different parts of the embryo (unpublished data), showing that DRN not only also has a function upstream of auxin, but also downstream. DRNL regulatory sequences also contain AREs analogous in position and number to those for DRN (Fig. 1A) and we postulate that these elements may also be functional. Since auxin is also synthesized at the tips of the developing cotyledons,2 in a similar domain to that of DRN expression, further work is needed to dissect the relevance of auxin transport from the regulation of biosynthesis.
Figure 1.
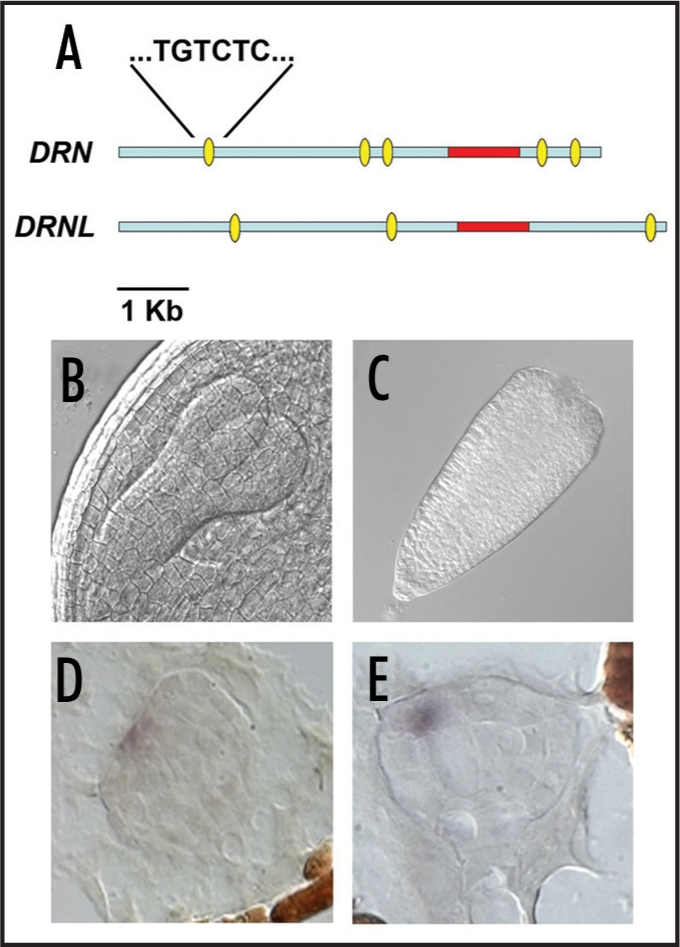
Comparison of the position of the TGTCC auxin response elements (ovals) within DRN and DRNL upstream and downstream genomic sequences. The DRN and DRNL coding regions are shown within this genomic region (A). Nomarski image of a wild type torpedo embryo (B) or a drn-1 drnl-2 double mutant embryo at the same developmental stage, showing the absence of cotyledons (C). RNA in situ hybridizaion of an STM probe on wild type (D) or drn-1 drnl-2 double mutnat (E) transition stage embryos, showing the misexpression of STM in the mutant background.
DRN/DRNL Affect Radial Symmetry
Cotyledon initiation demarcates the change from radial to bilateral symmetry during embryo development. Higher order mutants in auxin signalling or transport such as pin1 pin3 pin4 pin7 and pin1 pid result in cotyledonless embryos, a phenotype shared by the laterne mutant, a result of mutation in both PINOID and ENHANCER OF PINOID,1,15,16 underlining the importance of auxin during cotyledon ‘anlage’ initiation. drn drnl double mutants containing a strong drnl allele completely lack cotyledons, thereby abolishing radial symmetry (Fig. 1B and C). These double mutant embryos are a useful tool with which to study meristem development, since they initiate leaves, demonstrating firstly that STM function is not impaired, and secondly, that leaf initiation can be uncoupled from cotyledon formation. The study of other meristem-specific genes in drn-1 drnl-2 double mutants, such as SHOOTMERISTEMLESS (STM), showed that although STM expression is not affected by loss of DRN and DRNL function, its spatial expression domain is altered (Fig. 1D and E), showing that DRN and DRNL provide positional information and control radial symmetry. Current work aims to identify the direct downstream targets of DRN, to elucidate the transcriptional cascades involved in its function.
Redundancy May Reside in Higher Order Protein-Protein Complexes
DRN and DRNL proteins can physically interact in planta with PHAVOLUTA via their AP2 domains, and potentially with all members of the class III HD-ZIP family, via a novel C-terminal PAS-like domain. This potential multiplicity of protein-protein interactions for DRN and DRNL and the redundant control of embryonic patterning in general raises the possibility that the robustness of embryo development may at least partially reside in the promiscuity of protein-protein interactions, with specificity being conferred by particular combinations of higher order transcription factor complexes subsequently affecting auxin fluxes and response. Disruption in KANADI or HD-ZIP class III gene families responsible for normal axis specification, compromises polar auxin transport via PIN1,10 and supports this model. The identification of these higher order complexes and the transcriptional domains involved in their mutual function is a necessary goal in the challenge to dissect individual gene contributions within similar pathways.
Recent Insights into DRNL Function
DRNL also been recently published as SOB2,17 ESR218 and BOLITA,19 suggesting diverse functions in light responses, shoot regeneration, and cell expansion and proliferation. Microarray analysis has shown that DRNL over-expression results in the altered regulation of many auxin-responsive transcripts, which strengthens a proposed function for DRNL in auxin signalling pathways,19 and in the transcriptional regulation of CUC1.18 Additionally, DRNL function has been implicated in stamen development,20 suggesting a possible analogous function in organ boundary separation in flowers to that during cotyledon development.
Perspectives
The phylogenetic study of DRN homologues from other species addresses evolutionary developmental questions: expression of the putative DRN orthologue in maize, ZmDRN, pre-patterns the presumptive scutellum domain at the late proembryo stage,21 and lateral organ ‘anlagen’, as in Arabidopsis.22 As the scutellum is a grass-specific organ, thought to be homologous to the cotyledon structure of dicots, this, together with the fact that putative DRN orthologues have also been isolated for rice,21 suggests a conservation of DRN function across the monocot/dicot division.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/4864
References
- 1.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–152. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 2.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 3.Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development. 2001;128:4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
- 4.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes and auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aida M, Ishida T, Fukaki H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vroeman CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot apical meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibara K, Karim MR, Takada S, Taoka K, Rurutani M, Aida Tasaka M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18:2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clarke SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izhaki A, Bowman JL. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 2007;19:495–508. doi: 10.1105/tpc.106.047472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler JW, Cole M, Flier A, Grewe B, Werr W. The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development. 2007;134:1653–1662. doi: 10.1242/dev.001016. [DOI] [PubMed] [Google Scholar]
- 12.Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Reinhardt D, Pesce ER, Steiger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 14.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-responsive factors. Proc Nat Acad Sci USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furutani M, Vernoux T, Traas J, Kato T, Tasak M, Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidospis embryogenesis. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- 16.Treml BS, Winderl S, Radykewicz R, Herz M, Schweizer G, Hutzler P, Glawischnig E, Torres Ruiz RA. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development. 2005;132:4063–4074. doi: 10.1242/dev.01969. [DOI] [PubMed] [Google Scholar]
- 17.Ward J, Smith AM, Shah PK, Galanti SE, Yi H, Demianski AJ, van der Graaff E, Keller B, Neff MM. A new role for the Arabidopsis AP2 transcription factor LEAFY PETIOLE, in gibberellin-induced germination is revealed by misexpression of a homologous gene, SOB2/DRN-LIKE. Plant Cell. 2006;18:29–39. doi: 10.1105/tpc.105.036707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda Y, Banno H, Niu QW, Howell S, Chua NH. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006;47:1443–1456. doi: 10.1093/pcp/pcl023. [DOI] [PubMed] [Google Scholar]
- 19.Marsch-Martinez N, Greco R, Becker JD, Dixit S, Bergervoet JHW, Karaba A, de Folter S, Pereira A. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol Biol. 2006;62:825–843. doi: 10.1007/s11103-006-9059-1. [DOI] [PubMed] [Google Scholar]
- 20.Nag A, Yang Y, Jack T. DORNRÖSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Mol Biol. 2007;65:219–232. doi: 10.1007/s11103-007-9210-7. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann R, Werr W. Transcription of the putative maize orthologue of the Arabidopsis DORNROESCHEN gene marks early asymmetry in the proembryo and during leaf initiation in the shoot apical meristem. Gene Expr Patterns. 2007;7:158–164. doi: 10.1016/j.modgep.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Kirch T, Simon R, Grunewald M, Werr W. The DORNROESCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell. 2003;15:694–705. doi: 10.1105/tpc.009480. [DOI] [PMC free article] [PubMed] [Google Scholar]


