Abstract
The appropriate specification of distinct cell types is important for generating the proper tissues and bodies of multicellular organisms. In the root epidermis of Arabidopsis, cell fate determination is accomplished by a transcriptional regulatory circuit that is influenced by positional signaling. A leucine-rich repeat receptor-like kinase, SCRAMBLED (SCM), has been shown to be responsible for the position-dependent aspect of this epidermal pattern. In a recent report, we find that SCM affects the transcriptional regulatory network by down-regulating the WEREWOLF (WER) MYB gene expression in a set of epidermal cells located in a specific position. We also find that SCM and the SCM-related SRF1 and SRF3 are not required for embryonic epidermal patterning and that SRF1 and SRF3 do not act redundantly with SCM. This suggests that distinct positional signaling mechanisms exist for embryonic and post-embryonic epidermal patterning. In this addendum, we discuss the implications of our recent findings and extend our working model for epidermal cell pattering.
Key words: Arabidopsis, cell fate, leucine-rich repeat receptor-like kinase, pattern formation, positional signaling, root epidermis, root hairs
Introduction
Understanding the strategies that organisms use to define their cell fates and the ways that intrinsic and extrinsic cues regulate this process represent a major challenge in developmental biology. The specification of the epidermal cells of the Arabidopsis root provides a simple experimental system for defining the molecular basis of cell specification in plants. These cells have a binary cell-fate choice: root-hair cell or non-hair cell. Molecular genetic analyses suggest that a transcription complex including the WER-GL3/EGL3-TTG proteins (the active complex (AC)) defines the non-hair cell fate. This active complex is regulated bi-directionally by CPC and GL3 to ensure that adjacent epidermal cells adopt different fates.1 Further, epidermal cell fate is influenced by their location in relation to other cells, implying that positional signaling plays a role in pattern formation.2,3 Developing epidermal cells outside two underlying cortical cells (designated the H position) normally adopt root hair cell fate, whereas epidermal cells outside a single cortical cell (designated the N position) differentiate as non-hair cells. Recently, we showed that SCRAMBLED (SCM), a leucine-rich repeat receptor-like protein (LRR-RLK) is required for the proper interpretation of the positional information.4 In a paper appearing in the February 2007 issue of Developmental Biology, we uncovered new insights into the mechanistic and temporal aspects of SCM-mediated signaling in this model system.
Spontaneous Cell Fate Decisions in the Absence of Positional Signaling
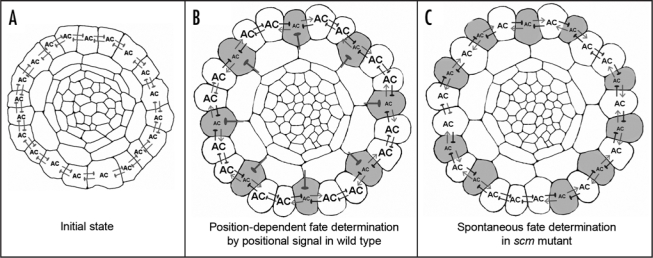
A detailed analysis of the cell type distribution in the scm mutants revealed that positional signaling is uncoupled from the lateral regulatory circuit. That is, we find a non-random distribution of hair cells and non-hair cells, but the location of these cells are no longer strictly determined by their position. This suggests that epidermal cells in the scm mutant are still communicating with each other through lateral regulatory circuit to adopt their fates (Fig. 1). Further, there is an appropriate correspondence in gene expression in the developing scm epidermal cells; for example, WER::GFP-expressing cells also express GL2::GUS4, and CPC::GUS, which are non-hair cell specific, but not EGL3::GUS which is root-hair-cell specific.5–7 Furthermore, loss of SCM function doesn't affect the distribution of the CPC-GFP protein,8 and only GL2::GUS expressing cells possess GL3-YFP protein (which preferentially accumulates in non-hair cells5) in their nucleus. Together, these results show that the SCM-mediated positional signaling acts independently of the CPC- and GL3-mediated lateral communication. The separate nature of these two cell signaling activities may provide an explanation for the evolutionary origin and the robustness of root epidermal cell specification.
Figure 1.
Pattern formation in the root epidermis of Arabidopsis. Schematic drawings of transverse root sections of (A) early stage, (B) late stage of wild-type, and (C) late stage of scm mutant. Shaded cells represent cells that adopt the hair cell fate. The thick blunted lines radiating from between cortical cells in panel B represents the SCM-mediated positional signal, which represses AC (the activator complex, which contains WER, GL3/EGL3, and TTG). The blunted lines between epidermal cells represent the mutual inhibition mediated by CPC. The arrows emanating from the developing hair cells in (B and C) represent the GL3/EGL3 signal.
SCM Negatively Regulates WER Expression
A central question that arose following the identification of the SCM receptor revolved around the target of the SCM signaling pathway. It was conceivable that this pathway could create a bias in the activity or level of any component in the active complex or the inactive complex. Several lines of evidence from our recent work suggest that the primary effect of SCM is to negatively regulate WER gene expression. In cpc-1 scm-2 double mutants, most of cells in H and N files express WER::GFP but the fluorescence intensity in each cell is not altered. Real time RT-PCR analysis indicates a two-fold increase in WER transcript level in this line. Further, over-expression of SCM by the WER promoter reduces WER::GFP expression and WER mRNA level. We envision that the negative effect of SCM action on WER expression should be sufficient to create an initial bias in WER level (and thereby the activation complex (AC)) which determines the direction of the lateral feed back loop amplifying the bias (Fig. 1).
SCM Functions During a Specific Developmental Stage
It is known that position-dependent patterning of epidermal cells is initiated during embryogenesis.9 However, SCM is not required for epidermal patterning in the embryo and hypocotyls. We tested the redundancy of the two most similar LRR-RLKs of SCM in the LRR-V subfamily, SRF1 and SRF3, but we found that these are not involved in epidermal patterning during any developmental stage. In a related finding, the aerial phenotypes of scm/strubbelig mutants are not altered by the srf1 or srf3 mutations or 35S::SRF1 or 35S::SRF3 lines,10,11 further suggesting that SCM and SRF1/SRF3 are unrelated functionally. Thus we believe that positional signaling might be operating quite differently in the embryonic protoderm. Indeed, it is likely that “residual” patterning from the embryo has an impact on the postembryonic root pattern in the scm mutant. It may be that daughter cells from a given embryonic cell have a tendency to adopt the same fate as the mother cell. In this view, an inappropriate cell fate would only be adopted after the dominating factors are diluted sufficiently through multiple rounds of division. Indeed, the analysis of rare epidermal clones in the scm mutant supports this view. These scm epidermal cell clones, which arise from perpendicular divisions of a single cell in the H position and generate a set of cells in the H position and in the N position,12 contain a higher proportion of misspecified cells than the root as a whole. This suggests that an embryonic patterning mechanism influences the postembryonic epidermal pattern. Further, in the scm mutant, a major fraction of the epidermal clones contain cells that adopt the same fate, which suggests that, over multiple rounds of division, the cells are unable to sense their location and fail to adopt distinct fates.
Conclusion and Perspectives
The mediating of positional signaling by SCM in epidermal patterning is a fascinating and useful model for studying the roles of receptor-like kinases in plant development. Our finding of a linkage between SCM and the epidermal transcriptional network provides new insight into the mechanism of cell fate decisions. In the future, it will be important to understand how SCM affects WER gene expression, the nature of the SCM ligand, and the mechanism of embryonic epidermal pattern formation.
Addendum to: The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Kwak S-H, Schiefelbein J. Developmental Biol. 2007;302:118–131.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: www.landesbioscience.com/journals/psb/article/4969
References
- 1.Schiefelbein J, Lee MM. A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis. Phyisol Plant. 2006;126:503–510. [Google Scholar]
- 2.Cormack RGH. Investigations on the development of root hairs. New Phytol. 1935;34:30–54. [Google Scholar]
- 3.Galway ME, Masucci JD, Lloyd AM, Walbout V, Davis RW, Schiefelbein J. The TTG gene is required to specify epidermal cell fate and cell patterning in Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 4.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 5.Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 6.Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 7.Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 8.Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, Sato S, Tabata S, Okada K, Wada T. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y, Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyls of Arabidopsis. Development. 2001;128:3697–3705. doi: 10.1242/dev.128.19.3697. [DOI] [PubMed] [Google Scholar]
- 10.Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K. STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:9074–9079. doi: 10.1073/pnas.0503526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyüboglu B, Pfister K, Haberer G, Chevalier D, Fuchs A, Mayer KFX, Schneitz K. Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol. 2007;7:16. doi: 10.1186/1471-2229-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger F, Hung CY, Dolan L, Schiefelbein J. Control of cell division in the root epidermis of Arabidopsis thaliana. Dev Biol. 1998;194:235–245. doi: 10.1006/dbio.1997.8813. [DOI] [PubMed] [Google Scholar]