Abstract
The nitrogen-fixing symbiosis between bacteria in the family Rhizobiaceae and members of the legume family (Fabaceae) has been well studied, particularly from the perspective of the early signaling and recognition events. Recent studies of non-nodulating legume mutants have resulted in the identification of a number of genes that are responsive to signal molecules from the bacteria. However, a second group of nodule-forming bacteria, completely unrelated to the Rhizobiaceae, which are α-Proteobacteria, has been discovered. These bacteria belong to the β-Proteobacteria and have been designated β-rhizobia to distinguish them from the better-known α-rhizobia. Here, we review what is known in this economically important symbiosis about the interaction between legumes and α-rhizobia, and we incorporate information, where known, about the β-rhizobia.
Key Words: biological nitrogen fixation, recognition, specificity, α-rhizobia, β-rhizobia
Introduction
Nitrogen (N) is an essential element for all known life forms to exist. Proteins and nucleic acids, which are the basic building blocks for the genetic material and proteins that facilitate all processes within a cell, including signaling, are N-based compounds. Although N is an abundant element in our environment, making up approximately 78% of the Earth's atmosphere as N2, the stable, covalent triple bond holding the N atoms together renders N ≡ N difficult to reduce and essentially non-utilizable by most organisms. Nevertheless, numerous nitrogen-fixing prokaryotes (diazotrophs) break this stable bond and reduce N ≡ N to ammonia. Examples include free-living bacteria such as Azotobacter, Clostridium and cyanobacteria as well as symbiotic bacteria that associate with plants, including rhizobia (α- and β-subclasses of Proteobacteria) and the actinomycete Frankia. This process is known as biological nitrogen fixation (BNF).
In this review, we will focus on the early stages-recognition and infection-in the Rhizobium-legume symbiosis. We will describe not only those bacteria classically known as rhizobia (Rhizobiaceae, α-subclass of Proteobacteria), but also those referred to as β-rhizobia (β-subclass of Proteobacteria). Investigating the interaction between β-rhizobia and legumes provides an opportunity not only to test whether the information accrued from studies of the α-rhizobia can be applied to β-rhizobia, but also to expand our knowledge of this important nitrogen-fixing symbiosis.
Prologue; Overview of the Rhizobium-Legume Symbiosis
The Rhizobium-legume symbiosis is a mutualistic relationship whereby Gram-negative, rod-shaped bacteria provide fixed nitrogen to a compatible host legume in exchange for a source of carbohydrates and a habitat, a novel root organ known as a nodule (Fig. 1). Three subfamilies exist in the legumes (family Fabaceae): Caesalpinioideae, Mimosoideae and Papilionoideae. Of the three, the majority of the genera in the Mimosoideae and Papilionoideae establish nitrogen-fixing root nodules with rhizobia, whereas in the most basal subfamily, the Caesalpinioideae, very few genera develop nodules.1 Two major groups of rhizobia have been recognized as being symbiotic partners in this interaction with legumes. One group consists of the α-rhizobia, including members of Rhizobiaceae (Mesorhizobium, Azorhizobium, Sinorhizobium, Bradyrhizobium, Rhizobium) as well as other α-Proteobacteria including Methylobacterium, which nodulates Lotononis and Crotolaria,2–4 Blastobacter denitrificans, which establishes nodules on Aeschynomene indica,5 and Devosia neptuniae, which nodulates Neptunia natans.6 The other major group is the β-rhizobia comprising Burkholderia, Cupriavidus (formerly Ralstonia)7 taiwanensis,8–12 and Herbaspirillum lusitanum.13
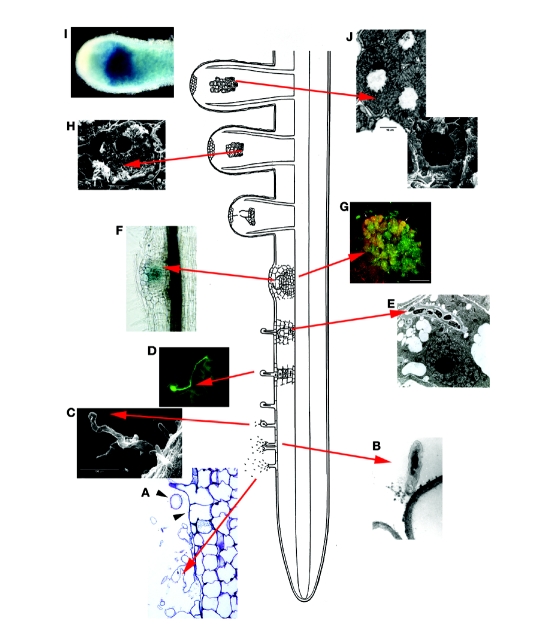
Figure 1.
Overview of the stages in the development of indeterminate nodules on plants, such as Medicago (papilionoid legume) or Mimosa (mimosoid legume). (A) Light microscopic (LM) view of the colonization of the root surface by rhizobia. The arrowheads point to polarly attached rhizobia. (B) Transmission electron micrograph (TEM) showing polar attachment of a rhizobial cell to a root hair. (C) Scanning electron micrograph (SEM) of the start of shepherd's crook formation. (D) LM of green-fluorescent protein (gfp)-labeled rhizobia within an infection thread with a shepherd's crook. (E) TEM of an infection thread within a developing nodule cell. (F) LM showing young nodule primordium; the rhizobia are labeled with gusA and stain blue. (G) Confocal laser scanning micrograph (CLSM) showing fluorescently labeled rhizobia within a young nodule. (H) SEM of a nodule cell containing elongating bacteroids. (I) A mature nodule containing lacZ-labeled rhizobia. (J) Two views (LM and SEM) of nitrogen-fixing bacteroids within a nodule cell. Modified from Niner and Hirsch.74
The family Rhizobiaceae is further subdivided into two large groups. The first encompasses those bacteria with a broad host-range, such as Bradyrhizobium, which is characterized by its ability to associate with several hosts. Another excellent example is NGR234, a Rhizobium strain that nodulates 232 species of legumes and even the non-legume, Parasponia andersonii.14 The second sub-group consists of the narrow host-range rhizobia, including Sinorhizobium and Mesorhizobium and other canonical α-rhizobial species, which are characterized by their ability to associate with only one or two closely related legume genera. The narrow-host range species, Rhizobium leguminosarum, is subcategorized into biovar-specific strains that correspond to the exact legume species nodulated. The acquisition of different symbiotic plasmids (pSym) in otherwise genetically identical chromosomal backgrounds gives the various R. leguminosarum biovars the ability to recognize their specific host.
Once nodules are formed on the plant roots and the bacteria successfully invade the root cells, the rhizobia differentiate into bacteroids, cells that undertake nitrogen fixation. The actual process of nitrogen fixation occurs through the action of nitrogenase, an oxygen-labile enzyme that facilitates the breakage of the dinitrogen bond. Both α- and β-rhizobia possess this enzyme, but in nodulating bacteria, it is only expressed under symbiotic conditions in the root nodule, where it can be protected from the damaging effects of oxygen.
The site of nitrogen and carbon exchange occurs at a membrane of plant origin called the peribacteroid membrane. This membrane envelops the differentiated bacteroid and together they form a functional nitrogen-fixing unit in the nodule known as the symbiosome. At this interface, the plant provides reduced carbon compounds to the bacteria and the bacteria supply fixed nitrogen to the host cell. This exchange is mediated by a series of membrane transporters and channels that are situated in the peribacteroid membrane, and is tightly regulated to maintain proper conditions for continued symbiosis.
Pas de Deux: Signaling and Recognition of the Partners
Establishing a complex relationship between two symbionts requires communication through the perception of signal molecules produced by each partner. This molecular “pas de deux” results in a coordinated series of events that culminates in the formation of a nodule. Two major molecules, one from the plant and the other synthesized by the rhizobia, which are involved in the earliest steps of symbiosis, are flavonoids and Nod factor. Flavonoids, a broad category of phenylpropanoid derivatives, are exuded by legume roots into the soil where they chemoattract bacterial cells to the root and also induce transcription of the rhizobial nod genes. The rhizobial nod gene products are utilized in the synthesis of Nod factor, a lipochitooligosaccharide (LCO) molecule, which is used for communicating with the host.15 Nod factor is composed of an oligomeric chain of three to five β-1,4-N-acetylglucosamine residues, with different substituents added to the backbone on either the reducing or non-reducing end. These chemical decorations provide specificity to the interaction. The regulatory gene nodD and the common nod genes, nodABC, whose gene products synthesize the LCO backbone, are on the large Sym (for symbiosis) plasmid or on the chromosome. They are found in all nodulating rhizobia, and are more or less interchangeable among rhizobial species. The host-specific nod genes are those required for interactions with a specific host species, and are not interchangeable among different strains of Rhizobium. They determine the variations in the substituents present on the Nod factor backbone. Once the plant roots perceive Nod factor, signal transduction cascades are triggered leading to nodule morphogenesis.
Divertissement: An Interlude with the β-Rhizobia
For over a hundred years, bacteria in the α-rhizobia and in particular the Rhizobiaceae were thought to be the only group of bacteria that could nodulate and fix nitrogen in association with legume roots. That idea radically changed when Moulin et al.16 demonstrated that members of the β-subclass of Proteobacteria inhabited legume root nodules. Since then, numerous reports have been published about both papilionoid and mimosoid legumes being nodulated by β-rhizobia.9–12,17–19 These bacteria have both nod and nif (for nitrogen fixation) genes, which are related to comparable genes in α-rhizobia, suggesting that lateral gene transfer has taken place, probably a long time ago.18 So far, nodA is the only nod gene that has been studied in detail for β-rhizobial phylogeny. NodA is an N-acyltransferase that mediates the transfer of an acyl group to the Nod factor backbone,20–21 and bona fide nodA homologs have not been found in bacteria other than rhizobia. However, nod gene sequencing in Burkholderia sp. STM678 (now known as B. tuberum)22 demonstrated that nodAB, preceded by a nodD-regulated nod box, are in a potential operon whereas an intact nodC is elsewhere in the genome.16 The organization of nod genes in C. taiwanensis is such that nodB and nodC are adjacent to each other and a nod box precedes them, but nodA is elsewhere in the genome.8 Both situations are contrary to what is typically found for Rhizobiaceae, although exceptions have been reported.23 Disruption of nodA resulted in a Nod− phenotype on the broad host range legume, Macroptilium atropurpureum,16 demonstrating that nodulation by Burkholderia is nod-gene dependent.
Although Nod factors have not been studied much in β-rhizobia, a chemical analysis has been undertaken of one from bacteria isolated from nodules of a number of Aspalathus species; these bacteria were originally identified as Bradyrhizobium aspalati. The LCOs are unsubstituted on the reducing end, but are highly substituted on the non-reducing end with fatty acids that vary in length and saturation, and are both N-methylated and 4,6 dicarbamoylated.24 Some α-rhizobia produce Nod factors that have similar substituents and also lack modified reducing ends.25 More investigations need to be pursued of Nod factors in the β-rhizobia to determine the extent of variation. For example, are host-specificity factors required? Which genes encode the host-specificity substituents (if any)? Detailing the differences in Nod factor structure between α- and β-rhizobia will help us determine whether the stages in Nod factor perception are also conserved (next section).
Terre-à-Terre: Legume Root Responses to Nod Factor
Legume roots respond to Nod factor at concentrations in the pico-molar range suggesting that a receptor with high affinity to Nod factor is involved in its perception. Once Nod factor is perceived, some responses occur very quickly, such as cellular calcium fluctuations.26 Some of the more protracted responses include cytoskeletal rearrangements,27 root hair deformation and curling (Fig. 1C and D),28 early nodulin gene expression and cell division leading to the inception of the nodule primordium (Fig. 1F and G).29 Nod factors also induce preinfection structures30 suggesting that they are involved in the initiation of infection threads. However, when added by themselves, they are unable to elicit full infection thread development in the root hairs.15
Allegro: Calcium Fluctuations
There are at least two phases of calcium responses in the root hairs: an initial influx of calcium (calcium flux) followed by an oscillation of calcium concentrations in the cytosol (calcium spiking). Calcium flux was first observed through measurements with micro-electrodes specific for certain ions.26,31 Upon Nod factor addition, a rapid (often within seconds) influx of calcium ions occurs, followed by an efflux of chloride and potassium ions, which results in alkalinization and depolarization of the cytoplasm, which damps out after about five minutes.32 These influxes were also measured by fluorescent dyes, which allows localized calcium fluxes to be visualized within root hairs. In these experiments, calcium gradients were found beginning with the highest concentration of calcium at the tip of the root hairs and tapering off towards the base of the root hair. Nod factor application enhanced this gradient and also induced a wave of increased calcium that travels from the tip of the root hair down the shaft. This phenomenon is found in a variety of legumes including Pisum sativum, Medicago sativa, Medicago truncatula and Phaseolus vulgaris, and appears to be related to root hair deformation.33–36
Calcium oscillations, cycling of calcium concentrations within the cell, are also observed in response to Nod factor application.33–38 The calcium spiking response takes longer to occur, at least three minutes or longer after Nod factor addition, and is characterized by a rapid increase in cytosolic calcium concentration followed by a slow decline. Calcium spiking can persist for up to an hour. The oscillations are mainly confined to the cytosolic regions surrounding the nucleus,33,35 and appear to be critical for nodule development based on the fact that legume mutants that do not undergo calcium spiking after Nod factor treatment are Nod−.37
Although there has not been any demonstration so far of the direct function of calcium fluctuations in inducing gene expression in nodulation, it has been shown in animal systems that calcium oscillations may lead to the regulation of gene transcription.39 Plants have been shown to respond to calcium oscillations by closing their stomata.40 Although the exact function of calcium fluctuations in nodulation is unknown, experiments with EGTA added to the external medium of Nod factor-treated plants suppressed both membrane depolarization and nodulin gene induction,31,41 providing further evidence that Nod factor signaling induces calcium oscillations needed for triggering downstream events that result in the development of nodules.
Adagio: Cytoskeletal Rearrangements and Root Hair Deformation
Actin microfilaments run along the length of the root hair from the tip to the base. Upon Nod factor application, these filaments begin to break down and accumulate at the root hair tip.27 Within minutes of Nod factor application, the root hair tips begin to bulge. Actin microfilaments at this stage are assembled at the cell membrane and grow in many directions, causing a bulge.42 As time passes, the microfilaments reassemble to cause polar growth of the root hair tip43 resulting in the beginnings of a deformed root hair. Cytochalasin (an inhibitor of actin polymerization)-treatment of Nod factor-induced root hairs prevented the polar growth of the root hair tip,42 supporting the idea that these types of cytoskeletal rearrangements are necessary for root hair deformation.
Root hair deformations come in a variety of forms. The initial response to Nod factor is the typical bulge or swelling at the tip of the root hair, followed by either a branching of the new root hair tip or twisting of the hair (Fig. 1A). Some of these deformations eventually produce a 360° curling of the root hair, known as the shepherd's crook, where the bacteria are trapped (Fig. 1C and D). It is within the tight curls that the bacteria initiate an infection thread to enter the cortical cells of the root (Fig. 1D and E). All of these morphological responses occur in the susceptible zone of the root, the site of rhizobial infection, where the root hairs begin to emerge (Fig. 1).
Perhaps surprisingly, Nod factor by itself or in combination with Nod-rhizobia does not elicit shepherd's crook formation or infection thread development on plants that develop indeterminate nodules (alfalfa, pea).30,44 On the other hand, Nod-mutants of certain broad host range rhizobia, which elicit determinate nodules (siratro, soybean), can be rescued by exogenous Nod factor.45,46 These differences have not yet been explained.
β-rhizobia also employ infection threads to enter the host roots upon which they initiate nodules. The threads appear to be initiated in root hairs based on studies on Mimosa species.10 Shepherd's crooks are formed and infection threads develop within the tightly curled hairs. The morphology of the Mimosa nodules is identical to those elicited by α-rhizobia (Fig. 1I),10 suggesting that nodule development follows the same program employed by the α-rhizobia. Table 1 summarizes the salient differences between the α- and β-rhizobia.
Table 1.
Comparisons between α- and β-rhizobia
| α-Rhizobia | β-Rhizobia | |
| Examples | Rhizobiaceae, Methylobacterium, Devosia neptuniae, Blastobacter denitrificans1 | Burkholderia species, Cupriavidus taiwanensis, Herbaspirillum lusitanum |
| nod genes | Common nod: nodABC and nodD regulatory gene; Host specificity nod: many different genes depending on species. | Common nod: nodABC and part of nodD. Others not yet identified. |
| Nod Factor | 3–5 glucosamine residues with a wide variety of reducing and non-reducing end substituents depending on the species. | 3–5 glucosamine residues with variable saturated and length fatty acids that are N-methylated and 4,6 dicarbamoylated; no reducing-end substituents identified so far. |
| Hosts | Mostly temperate papilionoid legumes | Subtropical to tropical mimosoid and papilionoid legumes |
van Berkum et al.73 have proposed changing Blastobacter denitrificans to Bradyrhizobium denitrificans.
Ballet D'Action: Nod Factor Receptors and Downstream Components
Earlier studies have shown that several single gene mutations in a legume host result in a Nod− phenotype. More recent investigations have focused on the identification of these genes. These studies have been facilitated by utilizing the model legumes Lotus japonicus (Lj) and Medicago truncatula (Mt).
The first gene to be cloned was LjNIN, for Nodule Inception, from L. japonicum;47 the P. sativum (pea) ortholog is PsSYM35. Mutants were characterized as being unable to form infection threads and hence lacked fully functional nodules. However, root hair curling is augmented compared to wild-type plants, indicating that the Nod factor perception machinery is intact in the mutants. A detailed analysis of the NIN gene indicated that the protein most likely functions as a transcriptional regulator of genes needed for nodulation, not only at inception but also later in nodule development.
Genes for other transcription factors have been recently identified in M. truncatula: NSP1 and NSP2, for Nodulation Signal Pathway genes. Genetic analysis indicates that these two genes encode proteins that are members of the GRAS family of transcription factors, and that NSP1 and NSP2 act downstream of the earliest signaling genes.48,49 NSP1 has been localized by GFP fusions to the nucleus of root hair cells,49 whereas NSP2 shows strong localization to the nuclear envelope and weak localization to the ER in spite of the lack of an ER retention signal.48 Upon Nod factor addition, the nuclear envelope localization is lost and fluorescence is detected in the ER and diffusely in the nucleus,48 whereas no change is observed for NSP1 localization upon adding Nod factor.49 This variation may partly reside in technical reasons because two different promoters were used to drive the GFP fusions.
The second gene to be cloned was the MsNORK (Nodule Receptor Kinase) gene in alfalfa (Medicago sativa), known as MtDMI2 in M. truncatula and PsSYM19 in P. sativum.50 Map-based positional cloning of the MsNORK gene identified it as encoding a transmembrane protein with several leucine-rich repeats and an intracellular region that contains serine/threonine kinase signatures.50 A comparable gene, called LjSYMRK for Symbiotic receptor kinase, was also identified in L. japonicus.51 The amino terminus of the nodule receptor kinase is unique to legumes, but a number of genes with limited similarity to NORK (NORK sequence-like or NSLs) exist, suggesting that this gene may be part of a large gene family in species as divergent as Arabidopsis.50 LRR domains are proposed to be involved in protein-protein interactions. Thus, MsNORK/MtDMI2 could interact with other components in the pathway, and subsequently transduce a signal via its serine/threonine kinase.
An interesting thing about Msnork/Mtdmi2 mutants is that they are also defective in mycorrhizal associations as are Mtdmi1 and Mtdmi3 mutants. The mycorrhizal symbiosis is a phosphate-acquiring mutualism in which more than 80% of land plants (but not Arabidopsis) engage in an interaction with mycorrhizal fungi. This symbiosis is believed to have originated more than 400 million years ago in contrast to the Rhizobium-legume symbiosis, which is thought to have evolved about 70 million years ago. Because of the ancient origin of the mycorrhizal symbiosis, it is believed that parts of the developmental pathway may have been coopted by the Rhizobium-legume symbiosis as evidenced by Mtdmi1, Mtdmi2 and Mtdmi3 mutants, which are both Nod− and Myc−. Also, the early nodulins ENOD2, ENOD12 and ENOD40 are expressed in mycorrhizal roots,52–53 further demonstrating the convergence of the mycorrhizal and nodulation pathways.
The predicted MtDMI1 protein is a transmembrane protein that is highly conserved among plant species. It contains a proline-rich region and a leucine zipper, which together make up a protein-protein interaction domain. The protein has similarity to an archael ligand-gated potassium channel,54 and hence may be a cation channel, but so far no data definitively show which ions are transported. The membrane depolarization and alkalinization responses that occur during calcium flux are not likely to be a consequence of MtDMI1 because dmi1 mutants still show calcium influx.36 MtDMI1 is associated with a membrane based on the transmembrane domain, but its exact location in the cell is not known for certain. Twin proteins, CASTOR and POLLUX, which are orthologs in L. japonicus and postulated to function as a heterodimer, carry chloroplast transit peptides and were shown by GFP fusions to be targeted to plastids.55 CASTOR and POLLUX are probably the result of a limited gene duplication event.55 Sequence comparisons show that POLLUX and MtDMI1 are more closely related than are CASTOR and MtDMI1. Whether the difference in one vs. two proteins depends on the formation of indeterminate vs. indeterminate nodules or just due to the limited examples available is unknown. More experiments need to be performed.
Cloning of the MtDMI3 gene (PsSYM9 in P. sativum) demonstrated its similarity to calcium/calmodulin-dependent serine/threonine protein kinases (CCaMKs).56 MtDMI3 resembles the calcium-dependent protein kinases in the N-terminal region in that they share similar kinases, but differs because of the presence of three calcium-binding EF hands rather than the typical four. Because Mtdmi3 mutants maintain the normal calcium flux and calcium oscillation responses, this protein most likely lies immediately downstream of these responses, and thus could use calcium as a second messenger to transduce the signal leading to early nodulin gene induction. Based on GFP fusion experiments, MtDMI3 localizes to the nucleus,48,49 although it lacks a canonical NLS based on our perusal of the protein sequence. Clearly, more studies need to be done.
Legume mutants that showed no calcium flux and were Nod− but Myc+ suggested that the mutated genes lie further upstream in the nodulation pathway. This led to the cloning of genes encoding potential Nod factor receptors. The most likely candidates are the LjNFR1 and LjNFR5 proteins from Lotus japonicus57,58 and their orthologs, PsSYM10 and PsSYM2A from pea, and MtNFP and MtLYK3/4 from M. truncatula.59 The predicted proteins have LysM domains and contain characteristics of receptor-like kinases. LysM domains are found in the E. coli MltD protein that binds the N-acetylglucosamine backbone of peptidoglycan, a component of the bacterial cell wall.60 In addition, LysM domains are also present in two proteins that bind chitin, which is chemically identical to the Nod factor backbone.61 Addition of chitin oligomers to legume roots activates calcium spiking similar to that observed upon addition of purified Nod factor.34 The existence of at least two proteins required for this stage of the symbiosis suggests that a small receptor complex could be involved in perception of Nod factor. Although these discoveries suggest an intriguing prospect for Nod factor receptors, the actual binding of Nod factor to LjNFR1/5 or other LysM-domain proteins has not been shown.
The calcium response of each of the mutants in the Nod factor signal transduction pathway is useful for placing these recently discovered genes within the nodule development pathway. Ljnfr1 and Ljnfr5 and its orthologs do not show any calcium responses, whereas Mtdmi1 and Mtdmi2 and their orthologs show a reduced calcium response, indicating that these genes are required for the characteristic calcium oscillations seen in response to Nod factor.62 In contrast, Mtdmi3 shows the full calcium response including calcium spiking, suggesting that this gene functions downstream of calcium signaling. It is not known whether mycorrhizal fungi also induce these types of calcium responses.
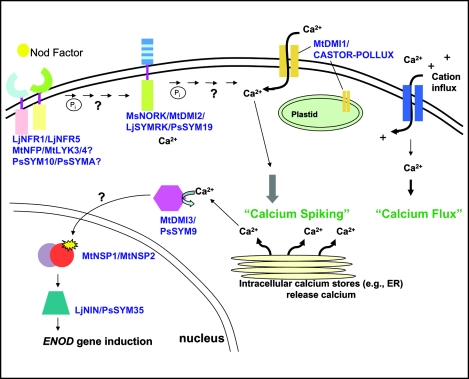
The binding of Nod factor to its putative receptor is proposed to activate transduction of the signal, presumably via phosphorylation of other proteins by a kinase domain. The model by which all these proteins fit into the early signaling events involved in Nod factor perception is illustrated in Figure 2. In this model, Nod factor binds to the receptor complex consisting of LjNFR1 and LjNFR5 or comparable LysM-domain proteins in other legumes (PsSYM10/PsSYM2A or MtNFP/MtLYS3/4). This complex could interact with MsNORK/MtDMI2/LjSYMRK/PsSYM19 nodule receptor kinases through their protein-protein interaction domains. Activation of these proteins leads to subsequent opening of an as-yet unidentified calcium channel as well as the cation channel encoded by MtDMI1/CASTOR-POLLUX, resulting in first, a rapid influx of calcium ions and second, calcium spiking. The exact localization of these various channels in the cell and their ion selectivity, however, will need to be resolved to determine whether these concepts are globally applicable. Activation of an anion channel, presumably in the plasma membrane, is also proposed to cause rapid membrane depolarization, but no mutants have been isolated for such a gene. Finally, increased cytosolic calcium levels through the action of MtDMI/CASTOR-POLLUX could activate calcium channels situated on the membranes of calcium stores (endoplasmic reticulum) to release calcium in a cyclic state (calcium oscillations). This could trigger the activation of MtDMI3/PsSYM9 (and transfer to the nucleus?) to phosphorylate and activate transcription factors such as NSP1 and NSP2 as well as LjNIN/PsSYM35 that are required for ENOD gene induction. Other reviews on this topic have been published and models proposed.62–65 No doubt, the models will be revised as more data become available.
Figure 2.
A likely pathway for Nod factor signaling in legumes based on the cloning of genes from various legume nodulation-defective mutants. Host-specific Nod factors produced by rhizobia are believed to interact with the LysM domains of Nod factor receptors such as LjNFR1 and LjNFR5. The involvement of the LysM domain-proteins in M. truncatula and P. sativum is not known and hence indicated by a question mark. Upon potentially being phosphorylated, one or both of the receptors is predicted to interact with a Nodule Receptor Kinase (MsNORK/MtDMI2/LjSYMRK/PsSYM19), triggering changes in membrane depolarization and ion flux through a potential cation channel and MtDMI1, which enables calcium spiking and oscillations. CASTOR and POLLUX, twin proteins in L. japonicus that are MtDMI ortho-logs, are localized to chloroplast membranes. Increased calcium levels from intracellular stores activate MtDMI3/PsSYM19, proteins highly similar to calcium/calmodulin protein kinase. The lack of an NLS, but experimental localization to the nucleus, suggests a possible transfer of DMI3 from the cytosol to the nucleus. This protein then triggers gene expression through MtNSP1 and MtNPS2 and LjNIN/PsSYM35, and other transcriptional regulators, which affect downstream gene expression.
Pas de Trois: α-, β-Rhizobia and Legumes
As more studies are being made of legumes, especially woody species that inhabit tropical and sub-tropical areas, more and more information is accumulating about nodulation and nitrogen fixation in these plants. Interestingly, the observation has been made that nodules containing β-rhizobia fix more nitrogen than α-rhizobia, in hosts that are nodulated by either group of nitrogen-fixing symbiont. For example, Mimosa pudica and M. pigra plants growing in Costa Rica and inoculated with Burkholderia and Cupriavidus spp. showed significantly higher biomass and acetylene reduction measurements than plants comparably inoculated with Rhizobium spp.19 Chen et al.10 made a similar observation regarding substantial nitrogenase activity for axenically grown Mimosa sp. inoculated with C. taiwanensis. In addition, many of the Mimosa-nodulating Burkholderia strains isolated by de Faria and colleagues in Brazil show high levels of nitrogen fixation and are used in soil reclamation.66 The reasons for the superior symbiotic parameters are unknown. It will be of interest to determine whether increased nitrogen fixation is a common trend for β-rhizobia and their hosts.
Another interesting observation is that B. cepacia, a human pathogen, which was isolated from nodules of Dalbergia louveli in Madagascar, was shown to induce efficient nodules on D. trichocarpa and Macroptilium atropurpureum. Surprisingly, nodA was not detected using either PCR or hybridization methods.67 The absence of a nodA gene may be due to extreme divergence between nod genes in B. cepacia and other nodulating burkholderias as suggested by Rasolomampianina et al.67 Interestingly, we have not been able to detect nodA in a Burkholderia isolated from the non-nodulating, but nitrogen-fixing caesalpinioid legume, Gleditsia triacanthos (Lum MR, de Faria SM, Hirsch AM, unpublished). If the absence of nodA is not uncommon, it argues that the paradigms established for legume interactions with α-rhizobia, which require nodA, may have to be modified to accommodate the differences between α- and β-rhizobia. So far, we know very little about legume-β-rhizobia interactions. Much of the current research is focused on discovery, but future research will have to address the issue of whether the models established for legumes and α-rhizobia are completely transferable to β-rhizobial interactions.
CODA: Conclusions and Perspectives
BNF results in a significant amount of nitrogen fixation per year, approximately 175 million metric tons of fixed nitrogen.68 Non-BNF reduced nitrogen, acquired from lightening or industrial processes, generates about 80 million metric tons of fixed nitrogen annually. Since its introduction in 1920, the Haber-Bosch process is responsible for increased food production and concomitantly much of the increase in the world's human population-from 2 billion then to the current more than 6 million.69 The Haber-Bosch process utilizes high heat (400–500°C) and pressure (200 atmospheres) to break the covalent N ≡ N bond to reduce nitrogen to ammonia for fertilizer. During the last half-century, the self-sufficiency and political stability of many countries are directly attributable to the Haber-Bosch process. However, our dependence on fossil fuels to generate the extreme conditions required to break the N ≡ N bond and the overuse of fertilizers in agriculture have frequently resulted in serious harm to the environment, including depletion of ozone and non-renewable resources (fossil fuels), pollution, an imbalance in the global nitrogen cycle and leaching of nitrate into groundwater.70 Recently, improved application of and use of lesser amounts of fertilizer have reduced some of the environmental damage of the last century. However, the use of petroleum-based fertilizers is likely to become unsustainable for modern-day food production, especially in non-oil producing countries, due to the rising costs of fuel.
By the year 2050, the world population is estimated to comprise 10.7 billion people, a nearly 80% increase in our current population, in spite of the fact that human fertility is starting to decrease.71 More food will have to be produced to keep pace with the additional mouths to feed. To accomplish this task, fertilizer production needs to be increased substantially. If not by means of the Haber-Bosch process, then how?
BNF so far is the only alternative to the Haber-Bosch process that provides sufficient fixed nitrogen, more than twice the amount produced by synthetic means, and does so in a sustainable manner. Basic research has provided us with a blueprint of how to proceed, but it will take a concerted effort to put basic knowledge into practical application. Two decades ago, we knew little of the genetic basis for nodule development and less than a decade ago, we were unfamiliar with the involvement of β-rhizobia in nodulation, nitrogen fixation and nitrogen economy. The β-rhizobia present another solution. The legumes they nodulate are indigenous to the sub-tropics and tropics, and thus contribute significantly to the nitrogen economy of these environments, many of which are in developing countries. Also, if it turns out that these bacteria are more efficient nitrogen-fixers than the α-rhizobia, it may be vital to engineer them (via modifications of their nod genes) to associate with agronomically important crop plants. Even if we are able to find alternative sources of energy, such as biofuels, for the Haber-Bosch process, it is unlikely that ample nitrogen will be fixed at the inexpensive prices of the past. Moreover, producing plants for nitrogen nutrition and human survival may overrule utilizing agricultural land for growing non-nitrogen fixing plants for sufficient biofuel manufacture. On the other hand, if high-yielding nitrogen-fixing plants are grown, two goals can be realized: food and biomass fuel for the future.
Like Haber in his acceptance speech for the Nobel Prize on June 2, 1920 for developing a synthetic means of nitrogen fixation, we must recognize the power of BNF and rethink our strategies for food production and security for the future.
Nitrogen bacteria teach us that Nature, with her sophisticated forms of the chemistry of living matter, still understands and utilizes methods which we do not as yet know how to imitate.72
Acknowledgements
This paper was written in partial fulfillment of the Ph.D. thesis of AL to the Department of Molecular, Cell and Developmental Biology, UCLA. AL was supported in part by UCBIOSTAR grant S98-86 and a grant from the Council of Research from the UCLA Academic Senate to AMH. NSF IOB-0537497 supports research on β-rhizobia in AMH's laboratory. We thank Drs. S.J. Kirchanski and M.R. Lum for their comments on the manuscript. We also thank the two anonymous reviewers for their helpful suggestions.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3143
References
- 1.Sprent JI. Nodulation in Legumes. Royal Botanic Gardens, Kew: The Cromwell Press Ltd.; 2001. p. 146. [Google Scholar]
- 2.Sy A, Giraud E, Samba R, de Lajudie P, Gillis M, Dreyfus B. Certaines légumineuses du genre Crotalaria sont spécifiquement nodulées par une nouvelle espèce de Methylobacterium. Can J Microbiol. 2001;47:503–508. (Fre). [PubMed] [Google Scholar]
- 3.Sy A, Giraud E, Jourand P, Garcia N, Willems A, de Lajudie P, Prin Y, Neyra M, Gillis M, Boivin-Masson C, Dreyfus B. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaftha JB, Strijdom BW, Steyn PL. Characterization of pigmented methylotrophic bacteria which nodulate Lotononis bainesii. Syst Appl Microbiol. 2002;25:440–449. doi: 10.1078/0723-2020-00124. [DOI] [PubMed] [Google Scholar]
- 5.Van Berkum P, Eardly BD. The aquatic budding bacterium Blastobacter denitrificans is a nitrogen-fixing symbiont of Aeschynomene indica. Appl Environ Microbiol. 2002;68:1132–1136. doi: 10.1128/AEM.68.3.1132-1136.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martinez-Molina E, Gillis M, Velazquez E. Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst Appl Microbiol. 2003;26:47–53. doi: 10.1078/072320203322337308. [DOI] [PubMed] [Google Scholar]
- 7.Vandamme P, Coenye T. Taxonomy of the genus Cupriavidus: A tale of lost and found. J Syst Evol Microbiol. 2004;54:2285–2289. doi: 10.1099/ijs.0.63247-0. [DOI] [PubMed] [Google Scholar]
- 8.Moulin L, Chen WM, Béna G, Dreyfus B, Boivin-Masson C. Rhizobia: The family is expanding. Nitrogen Fixation. In: Finan T, O'Brian M, Layzell D, Vessey K, Newton W, editors. Global Perspectives. New York: CABI Publishing; 2002. pp. 61–65. [Google Scholar]
- 9.Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Vandamme P. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol. 2001;51:1729–1735. doi: 10.1099/00207713-51-5-1729. [DOI] [PubMed] [Google Scholar]
- 10.Chen WM, James EK, Prescott AR, Kierans M, Sprent JI. Nodulation of Mimosa spp. by the β-Proteobacterium Ralstonia taiwanensis. Mol Plant-Microbe Inter. 2003;16:1051–1061. doi: 10.1094/MPMI.2003.16.12.1051. [DOI] [PubMed] [Google Scholar]
- 11.Chen WM, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C. Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J Bacteriol. 2003;185:7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett CF, Parker MA. Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst Appl Microbiol. 2005;28:57–65. doi: 10.1016/j.syapm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Valverde A, Velázquez E, Gutiérrez C, Cervantes E, Ventosa A, Igual JM. Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int J Syst Evol Microbiol. 2003;53:1979–1983. doi: 10.1099/ijs.0.02677-0. [DOI] [PubMed] [Google Scholar]
- 14.Pueppke SG, Broughton WJ. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant-Microbe Inter. 1999;12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 15.Long SR. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001;125:69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulin L, Mundive A, Dreyfus B, Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 17.Chen WM, James EK, Chou JH, Sheu SY, Yang SZ, Sprent JI. β-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. 2005;168:661–675. doi: 10.1111/j.1469-8137.2005.01533.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen WM, de Faria SM, Straliotto R, Pitard RM, Simões-Araùjo JL, Chou JS, Chou YJ, Barrios E, Prescott AR, Elliott GN, Sprent JI, Young JPW, James EK. Proof that Burkholderia strains form effective symbioses with legumes: A study of novel Mimosa-nodulating strains from South America. Appl Environ Microbiol. 2005;71:7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett CF, Parker MA. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environ Microbiol. 2006;72:1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkinson EM, Palcic MM, Hindsgaul O, Long SR. Biosynthesis of Rhizobium meliloti lipooligosaccharide Nod factors: NodA is required for an N-acyltransferase activity. Proc Nat Acad Sci USA. 1994;91:8418–8422. doi: 10.1073/pnas.91.18.8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Röhrig H, Schmidt J, Wieneke U, Kondorosi E, Barlier I, Schell J, John M. Biosynthesis of lipooligosaccharide nodulation factors: Rhizobium NodA protein is involved in N-acylation of the chitooligosaccharide backbone. Proc Nat Acad Sci USA. 1994;91:3122–3126. doi: 10.1073/pnas.91.8.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandamme P, Goris J, Chen WM, de Vos P, Willem A. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol. 2002;25:507–512. doi: 10.1078/07232020260517634. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XX, Turner SL, Guo XW, Yang HJ, Debellé F, Yang GP, Dénarié J, Young JPW, Li FD. The common nodulation genes of Astragalus sinicus rhizobia are conserved despite chromosomal diversity. Appl Environ Microbiol. 2000;66:2988–2995. doi: 10.1128/aem.66.7.2988-2995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boone CM, Olsthoorn MMA, Dakora FD, Spaink HP, Thomas-Oates JE. Structural characterisation of lipo-chitin oligosaccharides isolated from Bradyrhizobium aspalati, microsymbionts of commercially important South African legumes. Carb Res. 1999;317:155–163. doi: 10.1016/s0008-6215(99)00083-x. [DOI] [PubMed] [Google Scholar]
- 25.D'Haeze W, Holsters M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology. 2002;12:79R–105R. doi: 10.1093/glycob/12.6.79r. [DOI] [PubMed] [Google Scholar]
- 26.Felle HH, Kondorosi E, Kondorosi A, Schultze M. The role of ion fluxes in Nod factor signaling in Medicago sativa. Plant J. 1998;13:455–463. [Google Scholar]
- 27.Cárdenas L, Vidali L, Dominguez J, Perez H, Sanchez F, Hepler PK, Quinto C. Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 1998;116:871–877. doi: 10.1104/pp.116.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulfated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 29.Foucher F, Kondorosi E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol Biol. 2000;43:773–786. doi: 10.1023/a:1006405029600. [DOI] [PubMed] [Google Scholar]
- 30.Geurts R, Heidstra R, Hadri AE, Downie JA, Franssen H, van Kammen A, Bisseling T. Sym2 of pea is involved in a Nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol. 1997;115:351–359. doi: 10.1104/pp.115.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felle HH, Kondorosi E, Kondorosi A, Schultze M. Elevation of the cytosolic free Ca2+ is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol. 1999;121:273–279. doi: 10.1104/pp.121.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felle HH, Kondorosi E, Kondorosi A, Schultze M. Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 1996;10:295–301. [Google Scholar]
- 33.Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 34.Cárdenas L, Feijo JA, Kunkel JG, Sanchez F, Holdaway-Clarke T, Hepler PK, Quinto C. Rhizobium Nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 1999;19:347–352. doi: 10.1046/j.1365-313x.1999.00517.x. [DOI] [PubMed] [Google Scholar]
- 35.Walker SA, Viprey V, Downie JA. Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proc Nat Acad Sci USA. 2000;97:13413–13418. doi: 10.1073/pnas.230440097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw SL, Long SR. Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 2003;131:976–984. doi: 10.1104/pp.005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Dénarié J, Long SR. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Nat Acad Sci USA. 2000;97:13407–13412. doi: 10.1073/pnas.230439797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris JM, Wais R, Long SR. Rhizobium-induced calcium spiking in Lotus japonicus. Mol Plant-Microbe Inter. 2003;16:335–341. doi: 10.1094/MPMI.2003.16.4.335. [DOI] [PubMed] [Google Scholar]
- 39.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 40.Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffman T, Tang YY, Grill E, Schroeder JI. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature. 2001;411:1053–1057. doi: 10.1038/35082575. [DOI] [PubMed] [Google Scholar]
- 41.Pingret JL, Journet EP, Barker DG. Rhizobium Nod factor signaling: Evidence for a G protein-mediated transduction mechanism. Plant Cell. 1998;10:659–671. doi: 10.1105/tpc.10.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller DD, de Ruijter NCA, Bisseling T, Emons AMC. The role of actin in root hair morphogenesis: Studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 1999;17:141–154. [Google Scholar]
- 43.De Almeida Engler J, Ruijter NCA, Rook MB, Bisseling T, Emons AMC. Lipochito-oligosaccharides reinitiate root hair tip growth in Vicia sativa with high calcium and spectrin-like antigen at the tip. Plant J. 1998;13:341–350. [Google Scholar]
- 44.Hirsch AM, Asad S, Fang Y, Wycoff K, Löbler M. Molecular interactions during nodule development. In: Palacios R, Mora J, Newton WE, editors. New Horizons in Nitrogen Fixation. Dordrecht: Kluwer Academic Publishers; 1993. pp. 291–297. [Google Scholar]
- 45.Relic' B, Talmont F, Kopcinska J, Golinowski W, Promé JC, Broughton WJ. Biological activity of Rhizobium sp. NGR234 Nod-factors on Macroptilium atropurpureum. Mol Plant-Microbe Inter. 1993;6:764–774. doi: 10.1094/mpmi-6-764. [DOI] [PubMed] [Google Scholar]
- 46.Relic' B, Perret X, Estrada-García MT, Kopcinska J, Golinowski W, Krishnan HB, Pueppke SJ, Broughton WJ. Nod factors of Rhizobium are a key to the legume door. Mol Microbiol. 1994;13:171–178. doi: 10.1111/j.1365-2958.1994.tb00412.x. [DOI] [PubMed] [Google Scholar]
- 47.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 48.Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, Kiss GB, Downie, Oldroyd GED. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 49.Smit P, Raedts J, Portyanko V, Debellé, Gough C, Bisseling T, Guerts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 50.Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- 51.Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 52.van Rhijn P, Fang Y, Galili S, Shaul O, Atzmon N, Wininger S, Eshed Y, Lum M, Li Y, To V, Fujishige N, Kapulnik Y, Hirsch AM. Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc Nat Acad Sci USA. 1997;94:5467–5472. doi: 10.1073/pnas.94.10.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 1998;15:605–614. doi: 10.1046/j.1365-313x.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- 54.Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GED, Ayax C, Levy J, Debellé F, Baek JM, Kalo P, Rosenberg C, Roe BA, Long SR, Dénarié J, Cook DR. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303:1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- 55.Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, Pike J, Downie JA, Wang T, Sato S, Asamizu E, Tabata S, Yoshikawa M, Murooka Y, Wu GJ, Kawaguchi M, Kawasaki S, Parniske M, Hayashi M. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 2005;422:527–531. doi: 10.1038/nature03237. [DOI] [PubMed] [Google Scholar]
- 56.Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, Dénarié J, Rosenberg C, Debellé F. A putative Ca2+ and calmodul-independent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 57.Madsen EB, Madsen LH, Radutoiu S, Olbry M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J. A receptor kinase of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 58.Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 59.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 60.Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (Mltd) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 61.Butler AR, O'Donnell RW, Martin VJ, Gooday GW, Stark MJR. Kluyveromyces lactis toxin has an essential chitinase activity. Eur J Biochem. 1991;199:483–488. doi: 10.1111/j.1432-1033.1991.tb16147.x. [DOI] [PubMed] [Google Scholar]
- 62.Oldroyd GED, Downie JA. Calcium, kinases and nodulation signaling in legumes. Nat Rev Mol Cell Biol. 2004;5:566–576. doi: 10.1038/nrm1424. [DOI] [PubMed] [Google Scholar]
- 63.Geurts R, Fedorova E, Bisseling T. Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr Opin Plant Biol. 2005;8:346–352. doi: 10.1016/j.pbi.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 64.Stacey G, Libault M, Brechenmacher L, Wan J, May GD. Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol. 2006;9:110–121. doi: 10.1016/j.pbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Riely BK, Burns NJ, Ané JM. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbiosis. Genetics. 2006;172:2491–2499. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franco AA, de Faria SM. The contribution of N2-fixing tree legumes to land reclamation and sustainability in the tropics. Soil Biol Biochem. 1997;29:897–903. [Google Scholar]
- 67.Rasolomampianina R, Bailly X, Fetiarison R, Rabevohitra R, Béna G, Ramaroson L, Raherimandimy M, Moulin L, De Lajudie P, Dreyfus B. Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to α- and β-Proteobacteria. Mol Ecol. 2005;14:4135–4146. doi: 10.1111/j.1365-294X.2005.02730.x. [DOI] [PubMed] [Google Scholar]
- 68.Lynch JM, Hobbie JE. Microorganisms in Action: Concepts and Applications in Microbial Ecology. 2nd ed. Oxford, UK: Blackwell Scientific Publications; 1988. pp. 241–254. [Google Scholar]
- 69.Smil V. Enriching the Earth. Cambridge, MA: The MIT Press; 2001. p. 338. [Google Scholar]
- 70.Kinzing AP, Socolow RH. Human impact on the nitrogen cycle. Physics Today. 1994;47:24–35. [Google Scholar]
- 71.Smil V. How many billions to go? Nature. 1999;409:429. doi: 10.1038/46689. [DOI] [PubMed] [Google Scholar]
- 72.Haber F. Nobel Lecture. 1920. The synthesis of ammonia from its elements. ( http://nobelprize.org/chemistry/laureates/1918/haber-lecture.pdf) [Google Scholar]
- 73.Van Berkum P, Leibold JM, Eardly BD. Proposal for combining Bradyrhizobium spp. (Aeschynomene indica) with Blastobacter denitrificans and to transfer Blastobacter denitrificans (Hirsch and Muller, 1985) to the genus Bradyrhizobium as Bradyrhizobium denitrificans (comb. nov.) Syst Appl Microbiol. 2006;29:207–215. doi: 10.1016/j.syapm.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 74.Niner BM, Hirsch AM. How many Rhizobium genes, in addition to nod, nif/fix, and exo, are needed for nodule development and function? Symbiosis. 1998;24:51–102. [Google Scholar]