Abstract
Upon infection with necrotizing pathogens many plants develop an enhanced resistance to further pathogen attack also in the uninoculated organs. This type of enhanced resistance is referred to as systemic acquired resistance (SAR). In the SAR state, plants are primed (sensitized) to more quickly and more effectively activate defense responses the second time they encounter pathogen attack. Since SAR depends on the ability to access past experience, acquired disease resistance is a paradigm for the existence of a form of “plant memory”. Although the phenomenon has been known since the beginning of the 20th century, major progress in the understanding of SAR was made over the past sixteen years. This review covers the current knowledge of molecular, biochemical and physiological mechanisms that are associated with SAR.
Key Words: Arabidopsis; benzothiadiazole; defense response potentiation; 2,6-dichloroisonicotinic acid; elicitor; MAP kinase; parsley cell culture; priming; salicylic acid; sensitization
Introduction
In 1901, Beauverie and Ray independently realized that plants previously infected by a pathogen could better resist further infection.1,2 Over the 30 years that followed these reports, many studies suggested the existence of various induced disease resistance phenomena in plants. These have been summarized by Chester in 1933.3 One prominent induced resistance phenomenon is nowadays known as systemic acquired resistance (SAR).4–6 SAR is induced by most pathogens that cause tissue necrosis, either as a part of a hypersensitive response (HR)7 or as a symptom of disease.8 One characteristic of SAR is the development of the enhanced resistance in the distal, uninoculated plant organs.4–8 Another hallmark of SAR is its activity against a broad and distinctive spectrum of pathogens which includes viruses, bacteria, oomycetes, and fungi.4,9 In addition, SAR confers a long-lasting protection that can last for weeks to month, and sometimes throughout an entire season.9 Thus, for SAR to be realized, a plant requires the ability to recall past experience. Therefore, in addition to serving as a paradigm for signal transduction and having practical value, acquired resistance is a prime example for the existence of a form of “plant memory”.
Systemic SAR Signaling
Early grafting experiments have shown that a primary infected leaf of a plant can produce a systemic signal that is graft transmissible from rootstock to scion.10,11 The studies also revealed that the systemic signal induces SAR in the remote tissue in a species non-specific manner.10,11 The identity of the long-distance signal, however, remained unknown.4–6 Some experiments suggested salicylic acid (SA) was the translocated signal,12–15 but others argued against such a role for SA.16–18 Recent work with the Arabidopsis defective in induced resistance1 (dir1) mutant indicated that wild-type DIR1 with sequence similarity to lipid transfer proteins might play a role in the generation and/or transmission of a mobile signal for SAR, possibly by interaction with a lipid-derived molecule.19 Also, in the Arabidopsis enhanced disease susceptibility 1 (eds1) and phytoalexin deficient 4 (pad4) mutants, which are both defective in putative lipases,20,21 SAR cannot be activated.6 Together, these findings suggest that lipid signaling might contribute to SAR.
Hydrogen peroxide has also been proposed to have a signaling role in SAR.22 Inoculation of lower leaves of Arabidopsis plants with avirulent Pseudomonas syringae pv. tomato (Pst) DC3000 (avrRpt2) elicited a prominent H2O2 burst in the infected leaves. The oxidative burst was followed by the appearance of lower levels of H2O2 in small groups of cells in uninoculated leaves. The systemic “microbursts” were followed by the formation in the remote tissue of so-called “micro-HR” lesions. Using a pharmacological approach it has been shown that both the initial and secondary H2O2 bursts are both required for SAR.22 However, whether hydrogen peroxide might serve as a mobile systemic signal for SAR remained unclear.
Gaseous methyl salicylate, a major volatile produced in tobacco leaves inoculated with tobacco mosaic virus, was recently shown to function as an airborne signal which induces disease resistance in both the infected and non-infected tissues of an infected plant, and also in neighboring plants.23,24 Similarly, experiments with an ethylene-insensitive, transgenic tobacco line implicated ethylene as a signal for SAR, at least in tobacco.25 Together, the above findings indicate a complex nature for SAR long-distance signaling that might involve various different signals whose contribution to SAR might depend on the plant species.
Salicylic Acid: Endogenous Signal for SAR
Though the identity of the long-distance signal for SAR is unknown, it is appreciated that SA is needed to establish SAR in the remote tissue.4–6 First compelling evidence for this came from studies with transgenic tobacco and Arabidopsis plants constitutively expressing a bacterial SA hydroxylase. These plants are unable to accumulate high levels of SA and do not acquire systemic resistance upon infection with necrotizing pathogens,26,27 presumably due to the destruction of the SA signal. More recent work with Arabidopsis mutants affected in either SA production or SA signaling confirmed the role of SA as an important signal in SAR.28,29 In addition, in transgenic tobacco and Arabidopsis plants, overproduction of SA enhanced the resistance to pathogens.30,31
Chemical SAR Activators
Since it has been appreciated that SA is an endogenous signal for the activation of SAR, there was increased characterization of synthetic chemicals able to mimic SA in SAR induction. 2,6-dichloroisonicotinic acid and its methyl ester (both are referred to as INA) were the first synthetic compounds shown to activate SAR.32 As INA is insufficiently tolerated by some crops, benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) became an attractive synthetic SAR activator.33–35 SA, INA and BTH are assumed to activate SAR through a same signaling mechanism.4
SAR Genes
In tobacco and Arabidopsis, establishment of SAR is associated with the expression of a set of so-called SAR genes,36 which include some of those encoding pathogenesis-related (PR) proteins.37 Some PR proteins have been identified as acidic β-1,3-glucanases (BGL2) and chitinases (PR-3), possibly able to hydrolyze microbial cell wall components.37 Therefore, the accumulation of PR proteins has often been proposed as the molecular basis for SAR.
However, over the past few years it became widely appreciated that the accumulation of PR proteins does not per se explain the SAR phenomenon.38 For instance, cloning of PR genes and plant transformation by now have not provided a single example in which an inducible acidic glucanase or chitinase, alone or in combination, enhances resistance to fungal pathogens. Thus, the contribution of PR proteins to SAR appears to be minor.38
Priming (Sensitization)
Upon pathogen infection, there is activation of cellular defense responses in attacked cells of both susceptible and resistant plants. However, in case of (induced) resistance, cellular defense responses are induced more rapidly and stronger than in a susceptible interaction.9 Thus, an enhanced ability for the quick and effective activation of cellular defense responses that are induced not until challenge pathogen attack is another hallmark of SAR. According to the terminology for mammalian monocytes and macrophages the state of the enhanced ability to activate cellular defense responses was named the “primed” (“sensitized”) state of the plant (Fig. 1).9,39–41 Although the phenomenon has been known for years,9 priming and the resulting potentiation of cellular defense responses have not been widely appreciated until the early 1990s. This may, at least in part, be due to the fact that the enhanced ability of the tissue with SAR to better activate cellular defense responses does not become obvious until (challenge) pathogen attack of the protected tissue. In addition, the (enhanced) cellular defense responses are induced only in the few cells that are under attack, thus making their investigation difficult.
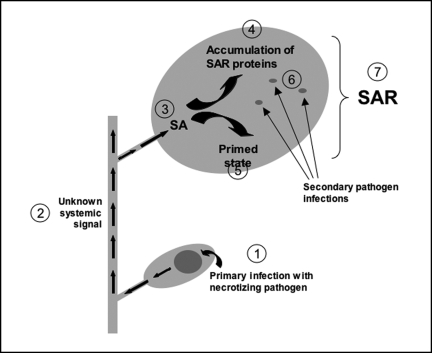
Figure 1.
Sequence of events associated with the establishment of SAR. Upon primary infection of a plant leaf with a necrotizing pathogen,1 a yet unknown systemic signal(s) is distributed systemically throughout the plant.2 The signal causes systemic accumulation of salicylic acid (SA).3 SA causes direct activation of SAR genes,4 some of which encode enzymes with antimicrobial activity. SA also conveys the tissue to the primed state5 which is characterized by an enhanced capacity to activate defense responses upon secondary pathogen attack. The faster and/or stronger activation of defense responses at the sites of secondary infection results in a decrease in disease symptoms,6 reflecting the SAR state.7
To overcome these difficulties and to provide a first systematic description of the priming phenomenon, the Kauss and Conrath laboratories took advantage of a parsley cell culture-elicitor model system to study priming and the resulting augmentation of cellular defense responses:40,41 Pretreatment with SA, INA or BTH, in a strictly time-dependent process, primed the parsley cells for strong activation already at low elicitor doses of various cellular defense responses.42–46 In more detailed studies with the parsley cell culture, Conrath and associates found that the effect of the SAR inducers on defense gene activation strongly depended on the gene that was monitored.45,46 One class of defense genes was found to be directly responsive to relatively low concentrations of SA or BTH.45,46 A second class of parsley defense-related genes was only slightly responsive to the treatment with any of the two SAR activators tested. Yet, already at low concentrations of SA or BTH, these genes displayed activator-dependent potentiation of their expression but only following treatment with a low dose of elicitor.45,46 For example, SA concentrations of 500 µM and higher were needed to activate the phenylalanine ammonia-lyase (PAL) defense gene using only SA, whereas as little as 10 µM SA greatly potentiated the subsequent elicitor-induced PAL expression.45 These results revealed a dual role for SAR inducers at the activation of plant defense responses: low doses of SA primed for potentiated activation of some defense genes, whereas higher doses directly activated other defense-related genes.
The first in-depth description of the priming phenomenon in whole plants was provided by Mur et al. in 1996.47 The authors reported that transgenic tobacco plants hydroponically pretreated with SA did not significantly express chimeric Asparagus officinalis PR-1::GUS or PAL-3::GUS defense genes. However, upon infection with P.s. pv. syringae or wounding, activation of the reporter genes was greatly enhanced.47 The SAR non-inducing SA analog 4-hydroxy benzoic acid was found inactive in this assay.47 Similar to the above mentioned results with systemically resistant tobacco plants, pretreatment with BTH was found to prime Arabidopsis for potentiated PAL expression upon infection with Pst DC3000.48 The BTH-induced priming also enhanced both PAL gene expression and callose deposition when these responses were induced by either mechanically wounding the leaves with forceps or by infiltrating them with water.48 The observations with Arabidopsis suggested that priming might be a common component that mediates cross-talk between pathogen defense, and wound and osmotic stress responses.48 Intriguingly, when SAR of Arabidopsis was biologically induced by previous infection with an avirulent strain of Pst (Pst DC3000 avrRpt2), there was potentiated activation also of PR-1 upon challenge infection with virulent Pst DC3000.48 Thus, it is likely that priming is the major mechanism in bona fide SAR, at least in Arabidopsis.
The Arabidopsis mutant enhanced disease resistance (edr1) shows constitutively enhanced resistance to Pst DC3000 and to the fungal pathogen Erisyphe cichoracearum.49 Interestingly, edr1 is different from other mutants with enhanced disease resistance because it shows no constitutive induction of PR-1 and BGL2, although transcripts of both genes accumulate in response to pathogen infection.49 This fact and the finding that edr1 displays stronger induction of defense responses such as the HR and callose deposition after infection strongly suggest an involvement of EDR1 in priming.
The Arabidopsis non-expresser of PR genes (npr1) mutant (npr1 also known as nim1 or sai1) accumulates wild-type SA levels in response to treatment with avirulent pathogens. However, the mutant is unable to express biologically or chemically induced SAR.50–52 Intriguingly, the potentiation by BTH-priming of both Pst-induced PAL expression and wound-or water infiltration-induced PAL gene activation and callose deposition are absent in npr1,48 strongly suggesting that a functional NPR1 gene is required for priming. Conversely, in the constitutive expresser of pr genes (cpr1) and cpr5 mutants of Arabidopsis, which express SAR in the absence of pretreatment with SAR activators,53,54 there was constitutive priming without BTH pretreatment, for potentiated PAL activation by Pst DC3000 infection, and for augmented induction of both PAL gene expression and callose deposition upon wounding or infiltrating the leaves with water.48 Though it cannot be excluded that constitutive priming in cpr1 and cpr5 might be due to the expression of a plethora of defense genes in these plants or due to activation of stress responses other than SAR, it is likely that because of the enhanced levels of SA in cpr1 and cpr5,53,54 these are permanently in the primed state. Because of constitutive priming, cpr1 and cpr5 might be able to rapidly and effectively induce their various cellular defense responses, thus leading to enhanced defense responses to pathogen attack, wounding or infiltration of water.4 It is noteworthy that the constitutively enhanced pathogen resistance of another Arabidopsis mutant, cpr5-2, has been ascribed to the potentiated induction of the PR-1 gene.55
The strong correlation between SAR and presence of priming, stressed the assumption that priming is a crucial mechanism in the bona fide SAR response of plants. The conclusion was further supported by the close correlation between the capability of various chemicals to induce SAR against tobacco mosaic virus in tobacco56 and their ability to prime for potentiated PAL expression induced by either elicitor treatment in parsley cells45,46 or Pst DC3000 infection, wounding or water infiltration in Arabidopsis plants.48 Moreover, an attenuation of priming and the concomitant loss of potentiated induction of the oxidative burst have been associated with a lack of resistance to avirulent bacterial pathogens in tobacco.57 And furthermore, overexpression in tomato of the disease resistance-associated gene PTI5 potentiates pathogen-induced defense gene expression and enhances the resistance to Pst.58
Observations similar to those made with parsley cell cultures, and tobacco, tomato, and Arabidopsis plants have been reported from SA-treated soybean cell cultures infected with P.s. pv glycinea,59 from BTH-primed and elicited Agastache rugosa suspension cells,60 and from SAR-induced and subsequently infected sunflower,61 cucumber,62 asparagus63,64 and cowpea plants.65 Together, these studies suggested that priming is a major mechanism of SAR in various different plant species.
Hypothetical Mechanisms of Priming: A Plant's “Memory”?
Induction of the primed state by SA, INA and BTH for augmentation of cellular defense responses was strictly dependent on an extended preincubation period.42,45,46 Therefore it has been hypothesized that the SAR inducers during pretreatment might cause time-dependent biosynthesis and preinfectional accumulation or posttranslational modification of cellular components with important roles in signal transduction and/or amplification. Accumulation or modification of the components per se would not activate most of the plant's defense responses. Yet due to their enhanced level in primed cells, there's augmented recruitment of the signal transmission components and, thus, potentiated activation of downstream defense responses but only upon subsequent exposure to biotic or abiotic stress.48 Recently, a mitogen-activated protein (MAP) kinase has been identified that accumulates upon priming in both parsley culture cells and Arabidopsis plants without displaying enzyme activity (Beckers, Zhang, Conrath, unpublished data). Probably due to the enhanced level of the inactive MAP kinase in primed cells and plants, there's augmented MAP kinase activity after stimulation by biotic or abiotic stress which is associated with enhanced induction of defense responses (Beckers, Zhang, Conrath, unpublished data). The priming-associated MAP kinase is a hot candidate for a cellular component that mediates priming.
Since various defense genes are activated faster and/or stronger in primed cells and plants than they are in nonprimed ones, the chromatin structure of defense-related genes has recently received increased attention when elucidating priming and SAR. As a matter of fact, the promoters of defense genes can be in more states than just “on” or “off” and the different stimuli that together cause promoter activation follow each other often with significant lag of time.66 In a recent pioneer study, Ng et al.67 investigated histone modifications associated with the inactive state, the primed state (inactive, but prepared for activation) and the active state of the β-phaseolin promoter in a heterologous Arabidopsis system. The studies uncovered that dependent on the promoter state, specific histone acetylation and methylation patterns were established in a specific manner indicating that a chromatin structure permissive for transcription can be established via progressive steps. In this scenario, an inactive promoter stores information in form of histone modifications to allow for a temporally separated transcriptional activation response. Whether similar processes play a role in priming for faster and/or stronger activation of defense genes, however, remains unclear. Interestingly, both systemic DNA rearrangements68 and alterations in the methylation pattern of DNA69 have been associated with the establishment of plant disease resistance.
NPR1: A Key Component for SAR
Over the past decade, a variety of mutants with compromised activation of SAR have been identified. The Arabidopsis mutant npr1 (see text above) is probably the most prominent of these mutants. Npr1 accumulates wild-type SA levels in response to infection with avirulent pathogens but is unable to activate PR genes,50–52 or establish the primed state,48 or develop biologically or chemically induced SAR.50–52 Thus NPR1 is a likely key regulator of SAR and priming. This assumption has further been supported by two studies demonstrating that constitutive overexpression of NPR1 in transgenic plants did not lead to enhanced SA levels or constitutive expression of PR genes. Rather, these plants showed stronger PR gene expression after pathogen infection and they also expressed greatly enhanced disease resistance.70,71 Interestingly, npr1 shows enhanced susceptibility to some virulent pathogens and seems to be involved also in R gene-mediated disease resistance.6 In addition, NPR1 seems to play a key role in the SA-independent induced systemic resistance (ISR) response.72 ISR is triggered by selected strains of saprophytic rhizobacteria and confers broad-spectrum disease resistance in the aerial parts of the plant.73 Impressively, in Arabidopsis activation of the NPR1-dependent ISR state is not associated with major changes in defense gene expression before pathogen infection.74 Rather, a plethora of defense-related genes shows augmented expression after pathogen attack,74 suggesting that NPR1-dependent priming is a major mechanism also in ISR.40,41 Similarly, the resistance that is induced by the non-protein amino acid β-aminobutyric acid (BABA) against Pst DC3000 and the fungal pathogen Botrytis cinerea in Arabidopsis also depends on an intact NPR1 gene.75,76 Intriguingly, the BABA-induced resistance to the pathogenic oomycete Hyaloperouospora parasitica was associated with a primed state enabling potentiated deposition of callose-containing papillae after infection.75
NPR1 has two domains likely mediating protein-protein interactions, an ankyrin repeat and a broad-complex, tramtrack, bric-á-brac/poxvirus, zinc finger (BTB/POZ) domain, as well as phosphorylation sites and a putative nuclear localization signal.6,77,78 Upon SAR activation, alterations in the redox state of the cell are the likely driving force that causes the inactive NPR1 homooligomer to convert to the active NPR1 monomer79 that accumulates in the nucleus80 where it interacts with a variety of proteins. These include members of the TGA family of basic leucine zipper transcription factors with a role in the activation of defense genes.81 A comprehensive literature review covering NPR1's role in SAR signaling has been published recently.6
The Costs of Priming and SAR
Many reports demonstrated that constitutive induction of disease resistance in plants might incur fitness costs. In most of these studies, defense responses have been activated by treatment of plants with high doses of SAR activators, transforming them with SA biosynthetic enzymes or by inducing gain-of-resistance mutations in Arabidopsis. For example, when wheat plants grown under pathogen-free conditions and under limited nitrogen supply were treated with BTH, the plants exhibited reduced growth and impaired seed set.82 Furthermore, transgenic Arabidopsis plants constitutively accumulating high levels of SA due to the expression of a SA synthase show a dwarfed phenotype and produce few seeds.31 In the cpr1, cpr5, and cpr6 mutants of Arabidopsis, which contain constitutively high levels of SA and permanent expression of PR genes,53,54 dwarfing has also been associated with reduced fitness of the plants.6
However, most of the studies aimed at evaluating the costs of resistance induction were based on the analysis of plants that had constitutively activated defense responses. The analyses did not consider that bona fide SAR is rather associated with the primed state for quicker and/or stronger activation of defense responses rather than with the direct activation of defense mechanisms. Recently, van Hulten et al.83 compared the costs of chemical priming by low doses of BABA with those for the direct induction of defense responses by high doses of BABA or BTH in Arabidopsis. The authors found that the direct induction of defense mechanisms seriously affected growth and seed set while priming had only marginal effects.83 Thus, priming seems to have economical advantages over the direct induction of the overall plant defense response.
Conclusion
SAR is a widely observed phenomenon in plants and priming for potentiated activation of defense responses has emerged as an important part of it (Fig. 1). In the primed state, plants are able to more quickly and more effectively activate defense responses when exposed to biotic or abiotic stress. A thorough elucidation of the primed state on the molecular, biochemical and physiological levels will not only contribute to a better understanding of signal transduction in plants. It will also enable utilization of the broad-spectrum, natural defense capacity of plants in the field. Finally, in-depth analysis of priming and SAR will provide access toward understanding this form of “plant memory”.
Acknowledgements
I apologize to my colleagues whose work could not be reviewed here because of space limitations. Work in my laboratory is funded by BASF, BASF Plant Science, Bayer and the Heinrich Hertz-Foundation.
Abbreviations
- BABA
β-aminobutyric acid
- BTH
benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester
- INA
2,6-dichloroisonicotinic acid (methyl ester)
- ISR
induced systemic resistance
- MAP kinase
mitogen-activated protein kinase
- NPR
non-expresser of PR genes
- PR
pathogenesis-related
- Pst
Pseudomonas syringae pv. tomato
- SA
salicylic acid
- SAR
systemic acquired resistance
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3221
References
- 1.Beauverie J. Essais d'immunisation des végétaux contre les maladies cryptogamiques. C R Acad Sci Ser III. 1901;133:107–110. (Fre). [Google Scholar]
- 2.Ray J. Les maladies cryptogamiques des végétaux. Rev Gen Bot. 1901;13:145–151. (Fre). [Google Scholar]
- 3.Chester KS. The problem of acquired physiological immunity in plants. Q Rev Biol. 1933;8:275–324. [Google Scholar]
- 4.Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sticher L, Mauch-Mani B, Métraux J-P. Systemic acquired resistance. Annu Rev Phytopathol. 1997;35:235–270. doi: 10.1146/annurev.phyto.35.1.235. [DOI] [PubMed] [Google Scholar]
- 6.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 7.Ross AF. Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology. 1961;14:329–339. doi: 10.1016/0042-6822(61)90318-x. [DOI] [PubMed] [Google Scholar]
- 8.Kuc J, Richmond S. Aspects of the protection of cucumber against Colletotrichum lagenarium by Colletotrichum lagenarium. Phytopathology. 1977;67:533–536. [Google Scholar]
- 9.Kuc J. Translocated signals for plant immunization. Annals New York Acad Sci. 1987;494:221–223. [Google Scholar]
- 10.Dean RA, Kuc J. Induced systemic protection in cucumbers: the source of the “signal”. Physiol Mol Plant Pathol. 1986;28:227–233. [Google Scholar]
- 11.Jenns AE, Kuc J. Graft transmission of systemic resistance of cucumber to anthracnose induced by Colletotrichum lagenarium and tobacco necrosis virus. Phytopathology. 1979;7:753–756. [Google Scholar]
- 12.Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 13.Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- 14.Shulaev V, León J, Raskin I. Is salicylic acid a translocated signal of systemic acquired resistance in tobacco? Plant Cell. 1995;7:1691–1701. doi: 10.1105/tpc.7.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mölders W, Buchala A, Métraux JP. Transport of salicylic acid in tobacco necrosis virus-infected cucumber plants. Plant Physiol. 1996;112:787–792. doi: 10.1104/pp.112.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen JB, Hamerschmidt R, Zook MN. Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 1991;97:1342–1347. doi: 10.1104/pp.97.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, Uknes S, Kessmann H, Ryals J. Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell. 1994;6:959–965. doi: 10.1105/tpc.6.7.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith-Becker J, Marois E, Huguet EJ, Midland SL, Sims JJ, Keen NT. Accumulation of salicylic acid and 4-hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-lyase activity in petioles and stems. Plant Physiol. 1998;116:231–238. doi: 10.1104/pp.116.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron R. A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- 20.Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;92:11312–11316. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez ME, Pennel RI, Meijer P-J, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 23.Shulaev V, Silverman P, Raskin I. Airborne signaling by methyl salicylate in plant pathogen resistance. Nature. 1997;385:718–721. [Google Scholar]
- 24.Seskar M, Shulaev V, Raskin I. Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 1998;116:387–392. [Google Scholar]
- 25.Verberne MC, Hoekstra J, Bol JF, Linthorst HJM. Signaling of systemic acquired resistance in tobacco depends on ethylene perception. Plant J. 2003;35:27–32. doi: 10.1046/j.1365-313x.2003.01778.x. [DOI] [PubMed] [Google Scholar]
- 26.Gaffney T, Friedrich L, Vernoij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 27.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1249. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 28.Dong X. Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol. 2001;4:309–314. doi: 10.1016/s1369-5266(00)00178-3. [DOI] [PubMed] [Google Scholar]
- 29.Van Wees SCM, Glazebrook J. Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv phaseolicola is due to degradation products of salicylic acid. Plant J. 2003;33:733–742. doi: 10.1046/j.1365-313x.2003.01665.x. [DOI] [PubMed] [Google Scholar]
- 30.Verberne MC, Verpoorte R, Bol JF, Mercado-Blanco J, Linthorst HJM. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nature Biotech. 2000;18:779–783. doi: 10.1038/77347. [DOI] [PubMed] [Google Scholar]
- 31.Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 2001;25:67–77. doi: 10.1046/j.1365-313x.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 32.Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, Uknes S, Ryals J. Induction of systemic acquired disease resistance in plants by chemicals. Annu Rev Phytopathol. 1994;32:439–459. doi: 10.1146/annurev.py.32.090194.002255. [DOI] [PubMed] [Google Scholar]
- 33.Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedrich L, Lawton K, Ruess W, Masner P, Specker N, Gut-Rella M, Meier B, Dincher S, Staub T, Uknes S, Métraux J-P, Kessmann H, Ryals J. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996;10:61–70. [Google Scholar]
- 35.Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- 36.Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux J-P, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- 38.Van Loon LC. Systemic induced resistance. In: Slusarenko AJ, Fraser RSS, Van Loon LC, editors. Mechanisms of Resistance to Plant Diseases. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 521–574. [Google Scholar]
- 39.Katz VA, Thulke OU, Conrath U. A benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol. 1998;117:1333–1339. doi: 10.1104/pp.117.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrath U, Pieterse CMJ, Mauch-Mani B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002;7:210–216. doi: 10.1016/s1360-1385(02)02244-6. [DOI] [PubMed] [Google Scholar]
- 41.Conrath U, et al. Prime-A-Plant Group Priming: getting ready for battle. Molec Plant-Microbe Interact. 2006 doi: 10.1094/MPMI-19-1062. in press. [DOI] [PubMed] [Google Scholar]
- 42.Kauss H, Theisinger-Hinkel E, Mindermann R, Conrath U. Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 1992;2:655–660. [Google Scholar]
- 43.Kauss H, Franke R, Krause K, Conrath U, Jeblick W, Grimmig B, Matern U. Conditioning of parsley (Petroselinum crispum) suspension cells increases elicitor-induced incorporation of cell wall phenolics. Plant Physiol. 1993;102:459–466. doi: 10.1104/pp.102.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kauss H, Jeblick W. Pretreatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiol. 1995;108:1171–1178. doi: 10.1104/pp.108.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thulke OU, Conrath U. Salicylic acid has a dual role in the activation of defense-related genes in parsley. Plant J. 1998;14:35–42. doi: 10.1046/j.1365-313X.1998.00093.x. [DOI] [PubMed] [Google Scholar]
- 46.Katz VA, Thulke OU, Conrath U. A benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol. 1998;117:1333–1339. doi: 10.1104/pp.117.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mur LAJ, Naylor G, Warner SAJ, Sugars JM, White RF, Draper J. Salicylic acid potentiates defense gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 1996;9:559–571. [Google Scholar]
- 48.Kohler A, Schwindling S, Conrath U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 2002;128:1046–1056. doi: 10.1104/pp.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frye CA, Innes RW. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10:947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;8:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah J, Tsui F, Klessig D. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the TMS2 gene. Mol Plant-Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 53.Bowling SA, Guo A, Cao H, Gordon AS, Klessig D, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant-Microbe Interact. 1998;12:1196–1206. [Google Scholar]
- 56.Conrath U, Chen Z, Ricigliano JR, Klessig DF. Two inducers of plant defense responses, 2,6-dichloroisonicotinic acid and salicylic acid, inhibit catalase activity in tobacco. Proc Natl Acad Sci USA. 1995;92:7143–7147. doi: 10.1073/pnas.92.16.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mur LAJ, Brown IR, Darby RM, Bestwick CS, Bi Y-M, Mansfield JW, Draper J. A loss of resistance to avirulent bacterial pathogens in tobacco is associated with the attenuation of a salicylic acid-potentiated oxidative burst. Plant J. 2000;23:609–621. doi: 10.1046/j.1365-313x.2000.00825.x. [DOI] [PubMed] [Google Scholar]
- 58.He P, Warren RF, Zhao T, Shan L, Zhu L, Tang X, Zhou J-M. Overexpression of PTI5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Molec Plant-Microbe Interact. 2001;14:1453–1457. doi: 10.1094/MPMI.2001.14.12.1453. [DOI] [PubMed] [Google Scholar]
- 59.Shirasu K, Nakajima H, Rajasekhar K, Dixon RA. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim HK, Oh S-R, Lee H-K, Huh H. Benzothiadiazole enhances the elicitation of rosmarinic acid production in a suspension culture of Agastache rugosa. Biotech Lett. 2001;23:55–60. [Google Scholar]
- 61.Prats E, Rubiales D, Jorrín J. Acibenzolar-methyl-induced resistance to sunflower rust (Puccinia helianthi) is associated with enhancement of coumarins on foliar surface. Physiol Molec Plant Pathol. 2002;60:155–162. [Google Scholar]
- 62.Cools HJ, Ishii H. Pretreatment of cucumber plants with acibenzolar-S-methyl systemically primes a phenylalanine ammonia-lyase (PAL1) for enhanced expression upon attack with a pathogenic fungus. Physiol Molec Plant Pathol. 2002;61:273–280. [Google Scholar]
- 63.He CY, Hsiang T, Wolyn DJ. Induction of systemic disease resistance and pathogen defense responses in Asparagus officinalis inoculated with non-pathogenic strains of Fusarium oxysporum. Plant Pathol. 2002;51:225–230. [Google Scholar]
- 64.He CY, Wolyn DJ. Potential role for salicylic acid in induced resistance of asparagus roots to Fusarium oxysporum f.sp. asparagi. Plant Pathol. 2005;54:227–232. [Google Scholar]
- 65.Latunde-Dada AO, Lucas JA. The plant defense activator acibenzolar-S-methyl primes cowpea [Vignia unguiculata (L.) Walp.] seedlings for rapid induction of resistance. Physiol Molec Plant Pathol. 2001;58:199–208. [Google Scholar]
- 66.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 67.Ng DWK, Chandrasekharan MB, Hall TC. Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell. 2005;18:119–132. doi: 10.1105/tpc.105.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 69.Kraska T. Vergleichende Untersuchungen von Resistenzinduktoren, deren Wirkungsweisen und dem Einfluß auf die DNA-Methylierung. Clausthal-Zellerfeld/Germany: Papierflieger; 1996. (Ger). PhD thesis, Hannover University. [Google Scholar]
- 70.Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friedrich L, Lawton K, Dietrich R, Willits M, Cade R, Ryals J. NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Molec Plant-Microbe Interac. 2001;14:1114–1124. doi: 10.1094/MPMI.2001.14.9.1114. [DOI] [PubMed] [Google Scholar]
- 72.Pieterse CMJ, van Wees S, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. A novel signaling pathway controling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Molec Plant-Microbe Interac. 2004;8:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 75.Zimmerli L, Jakab G, Métraux J-P, Mauch-Mani B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc Natl Acad Sci USA. 2000;7:12920–12925. doi: 10.1073/pnas.230416897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zimmerli L, Métraux J-P, Mauch-Mani B. β-aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Physiol. 2001;126:517–523. doi: 10.1104/pp.126.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H-Y, Johnson J, Delaney TP, Jesse T, Vos P, Uknes S. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 79.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 80.Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan W, Dong X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell. 2002;14:1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heil M, Hilpert A, Kaiser W, Linsenmair KE. Reduced growth and seed set following chemical induction of pathogen defense: Does systemic acquired resistance (SAR) incur allocation costs? J Ecol. 2000;88:645–654. [Google Scholar]
- 83.Van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]