Abstract
The basic structural unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around the histone octamer constituted by two molecules each of histones H2A, H2B, H3 and H4. Nucleosome assembly/disassembly/reassembly processes occur primarily during DNA replication and also during transcription, DNA repair and recombination. Several chromatin-remodeling factors had been previously shown to have pleiotropic roles in different processes of plant growth and development. We have recently demonstrated that the Arabidopsis NRP1 and NRP2 genes encode H2A/H2B chaperones and are required for the maintenance of post-embryonic root growth. The nrp1-1nrp2-1 double mutant plants specifically showed a short-root phenotype in normal growth conditions. They were also hypersensitive to DNA damage and showed release of transcriptional gene silencing. We propose that NRP1 and NRP2 act as histone H2A/H2B chaperones in nucleosome assembly, playing critical roles for a correct genome transcription in the maintenance of root growth.
Key Words: Arabidopsis thaliana, histone chaperone, chromatin, transcription, root growth
In plants as in animals, development is regulated by differential gene expression whereby cells acquire specific fates.1 Plant development takes place essentially post-embryonically. During embryogenesis, only the basic body plan is established, with small groups of cells called apical meristems at both ends of the body axis. During post-embryonic development, the shoot apical meristem (SAM) at the apical end and the root apical meristem (RAM) at the basal end continuously provide cells to maintain stem cells as well as to initiate organogenesis which leads to the development of the aerial parts and the subterranean root system of the plant, respectively. Chromatin structure is fundamental to transcription. Similar chromatin gene mutations that cause embryonic lethality in animals frequently result in plants that are detectably modified but viable, making it relatively easy to study the effects of these genes in development in plants. Chromatin-remodeling factors have been shown to play crucial roles in diverse processes of plant development, including leaf patterning, shoot organogenesis, flowering time control, gametogenesis and seed reproduction.2–5
By reverse genetic analysis, we demonstrated that the nrp1-1nrp2-1 mutant containing loss-of-function of both NRP1 and NRP2 genes has a short-root phenotype. NRP1 and NRP2 proteins show highest homology with the animal SET/TAF-I/I2PP2A proteins, which are involved in nucleosome assembly by chaperoning histones H2A and H2B.6 Consistently, NRP1 and NRP2 were found to bind histones, preferentially H2A and H2B than H3. They are localized primarily in the nucleus and they bind chromatin in planta. Additional supports of NRP1 and NRP2 in chromatin remodeling were the observations that the nrp1-1nrp2-1 mutant plants exhibit perturbed genome transcription, release of heterochromatic gene silencing and hypersensitive response to DNA damage.
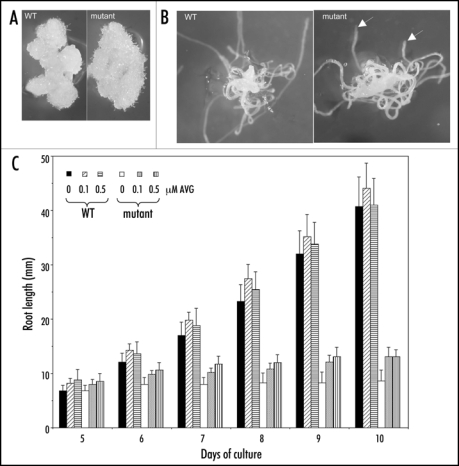
In spite of the fundamental role of NRP1 and NRP2 in chromatin remodeling, the phenotype of the nrp1-1nrp2-1 mutant is remarkably root specific, the embryos and the aerial organs (leaves, rosettes, inflorescences, flowers and fruits) developed normally in the mutant plants. Arrest of cell cycle progression at G2/M and disordered cellular organization were observed in the mutant roots. In this addendum, we show that the mutant root segments, when cultured in vitro, had similar capacity in callus formation than the wild-type root segments (Fig. 1A), and that roots regenerated in vitro from hypocotyls were also arrested in elongation in the mutant but not in the wild-type (Fig. 1B). These new data further strengthen the root specificity of requirement of NRP1 and NRP2 in the maintenance of cell proliferation. The specific short-root phenotype of the nrp1-1nrp2-1 mutant is in sharp contrast to the pleiotropic phenotypes of the fas1 and fas2 mutants, which include stem fasciation, abnormal phyllotaxy, modified leaf shape, reduced growth and size of all organs.7,8 FAS1 and FAS2 encode subunits of the CAF1 complex, which chaperones histones H3 and H4 in nucleosome assembly. It is possible that H2A/H2B and H3/H4 contribute to different levels of nucleosome assembly/disassembly and more importantly additional chaperones are likely involved in chaperoning histones in nucleosome assembly in Arabidopsis.
Figure 1.
Comparison of callus and root growth between the wild-type (WT) and the nrp1-1nrp2-1 mutant. (A) Representatives of callus regeneration and growth from root segments cultured in the presence of 2.3 µM 2,4-D, 11.4 µM IAA and 3.2 µM BAP. (B) Representatives of root regeneration and growth from hypocotyls cultured in the presence of 5.7 µM IAA and 1 µM IBA. Arrows indicate roots arrested in growth. Photographs were made two weeks after culture. (C) Root elongation of plants grown at different concentrations of the ethylene biosynthesis inhibitor AVG. The mean value from 20 plants is shown. Vertical bars represent standard deviations.
Several ethylene-responsive genes encoding transcription factors were upregulated in the nrp1-1nrp2-1 mutant seedlings. However, the mutant seedlings showed normal ethylene triple response (data not shown) and treatment by the ethylene biosynthesis inhibitor AVG (L-a-(2-amino-ethoxyvinyl)-glycine) could not sufficiently rescue the short-root phenotype (Fig. 1C). These latter observations indicate that the modifications of ethylene-responsive genes are not enough to explain the nrp1-1nrp2-1 mutant phenotype. Among the other differentially expressed genes found in the nrp1-1nrp2-1 mutant seedlings, GLABRA2 (GL2) was downregulated whereas PLETHORA2 (PLT2) was upregulated. GL2 encodes a homeodomain transcription factor and represses root hair formation,9 its downregulation correlated with the high proliferation of root hairs in the nrp1-1nrp2-1 mutant. PLT2 encodes an AP2-type transcription factor and plays crucial roles in stem cell specification and maintenance in the RAM.10 its perturbed expression could have significantly contributed to the nrp1-1nrp2-1 mutant root phenotype. NRP1 and NRP2 proteins were found to bind chromatin at GL2 and PLT2, thus might regulate directly expression of these genes. Chromatin organization at GL2 was also affected in fas2 mutant roots.11 In addition to dynamics of nucleosome assembly/disassembly, histone acetylation/deacetylation and ATP-dependent chromatin remodeling also play important roles in root growth and development.12,13 Future experiments will further explore epigenetic regulation in root growth and development, particularly in response to intrinsic and environmental factors.
NRP1 and NRP2 are expressed not only in roots but also in leaves, stems and flowers. In addition, NRP1 and NRP2 can form homomeric and heteromeric protein complexes, and NRP1 and NRP2 show functional redundancy in root growth. Nonetheless, molecular phenotype differed between the nrp1-1 and nrp2-1 single mutants: a significantly higher number of genes was detected by transcriptome analysis to be differentially expressed in the nrp2-1 than in the nrp1-1 mutant seedlings. Further work is required to understand both redundant and unique molecular functions of NRP1 and NRP2. The close homologues of NRP1 and NRP2 are NAP1-group proteins, which include four members (AtNAP1;1 to AtNAP1;4) in Arabidopsis. Different members of the NAP1-group proteins in rice and tobacco have distinct subcellular localizations and show different affinity to different types of histones.14,15 Genetic and molecular characterization of this group of genes in Arabidopsis will help to understand their roles in epigenetic inheritance, particularly interesting in comparison with NRP1 and NRP2 in plant growth and development.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3687
References
- 1.Meyerowitz EM. Plants compared to animals: The broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- 2.Guyomarc'h S, Bertrand C, Delarue M, Zhou DX. Regulation of meristem activity by chromatin remodelling. Trends Plant Sci. 2005;10:332–338. doi: 10.1016/j.tplants.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh TF, Fischer RL. Biology of chromatin dynamics. Annu Rev Plant Biol. 2005;56:327–351. doi: 10.1146/annurev.arplant.56.032604.144118. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- 5.Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Park YJ, Luger K. Structure and function of nucleosome assembly proteins. Biochem Cell Biol. 2006;84:549–558. doi: 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- 7.Leyser HMO, Furner IJ. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- 8.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science. 2003;300:1427–1430. doi: 10.1126/science.1083695. [DOI] [PubMed] [Google Scholar]
- 10.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Costa S, Shaw P. Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature. 2006;439:493–496. doi: 10.1038/nature04269. [DOI] [PubMed] [Google Scholar]
- 12.Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc Natl Acad Sci USA. 2005;102:14469–14474. doi: 10.1073/pnas.0503143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukaki H, Taniguchi N, Tasaka M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 2006;48:380–389. doi: 10.1111/j.1365-313X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 14.Dong A, Zhu Y, Yu Y, Cao K, Sun C, Shen WH. Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta. 2003;216:561–570. doi: 10.1007/s00425-002-0910-6. [DOI] [PubMed] [Google Scholar]
- 15.Dong A, Liu Z, Zhu Y, Yu F, Li Z, Cao K, Shen WH. Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 2005;138:1446–1456. doi: 10.1104/pp.105.060509. [DOI] [PMC free article] [PubMed] [Google Scholar]