Abstract
Nearly 240 WD repeat proteins have been identified from the Arabidopsis genome. Among these, some well characterized WDR proteins were shown to regulate various developmental processes in plants.1 We have recently isolated in Solanum chacoense a homolog of the Drosophila NOTCHLESS gene. In Drosophila, NOTCHLESS regulates the activity of the Notch signaling pathway through a direct interaction with the intracellular domain of the Notch receptor. Although the Notch signaling pathway does not exist in yease and plants, the NLE gene is conserved in animals, plants and yeast. Furthermore, functional conservation was suggested by expression of the plant NLE gene in Drosophila. In plants, underexpression of the plant NLE gene altered numerous developmental processes including seed development, and resulted in reduced aerial organ size and organ numbers, in delayed flowering, and in an increased stomatal index. Surprisingly, the link between these pleiotropic phenotypes is the recently discovered of the involvement of NLE in ribosome biogenesis, emphasizing its role in proper cellular growth and proliferation during plant development.
Key Words: notchless, notch, WD repeat protein, ovule, fertilization, ribosomal protein, ribosomal trans-acting factor, ribosome biogenesis
Notchless, a WD Repeat Protein Involved in a Variety of Developmental Pathways in Plants and Animals
Double-fertilization represents a pivotal event in the life cycle of angiosperms as it initiates the transformation of the ovule into a seed, a structure that carries the embryo of the next sporophytic generation. Following double-fertilization, a series of coordinated developmental processes are activated. One sperm cell fuses with the egg-cell to form the zygote and initiate embryogenesis, while another sperm cell unites with the central cell to initiate the development of an embryo-nourishing endosperm. Simultaneously, the integument(s) of the ovule differentiate into a protective seed coat while the ovary wall, within which the ovule is enclosed, differentiates into the fruit pericarp. Each of these developmental processes is linked to distinct genetic programs and, upon double-fertilization, their initiation involves major changes in gene expression.2–4
The ribosome is a key component in the machinery responsible for protein synthesis and, as such, plays a major role in controlling growth and development. The small 40S and large 60S subunits of the eukaryotic ribosome are made up of a total of four ribosomal RNAs (rRNAs) and about 80 ribosomal proteins (r-proteins). Unlike the prokaryotic ribosome, which is able to self-assemble, the biosynthesis of eukaryotic ribosomal subunits is highly coordinated in time and in space (from the nucleolus to the cytoplasm) and requires the participation of at least 170 nonribosomal factors.5 Nonribosomal factors and r-proteins are highly conserved in eukaryotes but, in contrast to other model organisms, limited information on their plant counterparts is available.6,7 Their importance during plant growth and development has however been shown by the expression of several plant r-protein genes being developmentally and environmentally regulated.8–12 Plant r-protein types are likely all encoded by small gene families6 and members of a r-protein gene family can be differentially regulated.12–14 Disruption of r-protein gene expression was shown to cause general growth delay,15–17 hypersensitivity to genotoxic stress18 and early embryonic developmental arrest.19 Information on plant nonribosomal factors is scarce, although one was reported to be essential for female gametogenesis.20
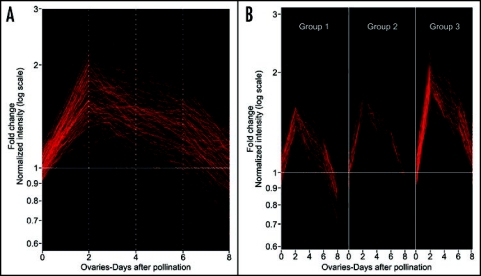
We recently reported that the NOTCHLESS (NLE) gene encodes a homolog of a yeast nonribosomal factor (YCR072, RSA4) and further provided evidences for an involvement of plant ScNLE protein in ribosome biogenesis by using yeast two-hybrid assays.21,22 In Solanum chacoense, transgenic plants with reduced levels of ScNLE transcripts displayed pleiotropic phenotypes including a severe reduction in seed set.23 Consistent with these phenotypes, we showed that ScNLE expression is associated to actively growing tissues of the shoot apex and is also strongly and transiently increased in ovules around fertilization time.21,23 In order to determine if the expression pattern of the ScNLE nonribosomal factor gene is shared by r-protein genes, we analyzed in this study the temporal expression profile of 288 ESTs corresponding to 65 r-proteins in ovules at different stages after fertilization by microarray analysis. DNA microarrays were performed from 7741 expressed sequence tags (ESTs) corresponding to 6374 ovule-expressed unigenes covering the zygotic to late torpedo embryonic developmental stages.2 Because ESTs were only partially sequenced, we grouped the r-protein gene sequences on the basis of their amino acid sequence similarity to r-protein types (Table 1) and not to individual members of a r-protein type gene family.6 Thus from our EST set, more than 80% of all the cytoplasmic r-protein types were represented. Interestingly, our DNA microarray analysis revealed that gene expression from all the r-protein genes analyzed peaked two days after pollination (DAP), similarly to the ScNLE nonribosomal factor gene.23 By 8 DAP, most had decreased to their basal level or below their level observed in unfertilized ovules (0 DAP). Self organization map (SOM) clustering revealed three distinct profiles (group 1 to 3). Most r-protein genes from group 1 showed a modest 1.5 to 2-fold increase in transcript levels 2 DAP and then decreased below the unfertilized ovule levels by 8 DAP. Most r-protein genes from group 2 showed a maximal 2-fold induction 2 DAP, and then returned to expression levels found in unfertilized ovules by 8 DAP. Most r-protein genes from group 3 showed greater than 2-fold increase in mRNA levels and a slower decrease back to levels found in unfertilized ovules.
Table 1.
Ribosomal proteins clustering with the self organizing map (SOM) groups as determined by cDNA microarray analysis
| Group 1 | Group 2 | Group 3 | |||
| SSU | LSU | SSU | LSU | SSU | LSU |
| S3 | L4 | S8 | L1 | S16 | L5 |
| S4 | L6 | S9 | L2 | S17 | L7a |
| S5 | L11 | S21 | L8 | S18 | L9 |
| S6 | L21 | S28 | L10a | S23 | L13 |
| S10 | L24 | L12 | S26 | L13a | |
| S11 | L32 | L34 | S27 | L14 | |
| S13 | L36 | L37 | S30 | L17 | |
| S14 | L38 | L37a | L18 | ||
| S15 | L41 | L22 | |||
| S19 | RAP* | L23a | |||
| S20 | P0 | L26 | |||
| S25 | P1 | L28 | |||
| S29 | P2 | L29 | |||
| S29a | P3 | L30 | |||
| Sa (P40) | L31 | ||||
| L33 | |||||
| L35 |
RAP, ribosomal associated proteins. Abbreviations: SSU, ribosome small subunit; LSU, ribosome large subunit.
Similarly to our observations, a significant increase in expression levels of several r-protein genes was observed in the zygote13 and two-celled proembryo.4 These data therefore suggest a rapid de novo transcription of r-protein genes upon fertilization as well as an increase in ribosome biogenesis and protein synthesis requirement at the onset of seed development. Our microarray analysis moreover showed a concerted upregulation of all the r-protein genes analyzed. Although members of a r-protein type family were shown to be differentially regulated, expression of most members seems to be upregulated when high supply of proteins is required, such as in actively growing and differentiating tissues.12,14 Since ribosome biogenesis depends on nonribosomal factors in eukaryotes,5 a similar transient upregulation profile in response to fertilization is expected for other plant nonribosomal factor encoding genes, as we previously demonstrated for the ScNLE gene.23 Interestingly, in animals, the NLE homolog was shown to interact with the intracellular domain of the Notch transmembrane receptor and to be a modifier of Notch activity by an unknown mechanism.24,25 The Notch signaling pathway is however metazoan-specific and, with the exception of NLE, components and regulators of this pathway have no homologs in yeast and plant genomes.26 Since the NLE gene and ribosome biogenesis as a whole are highly conserved in eukaryotes,7 this suggest that NLE is primarily involved in ribosome biogenesis and was likely later recruited as a regulator of the Notch pathway in the animal lineage. This raises the possibility that the Notch pathway is somehow regulated by ribosome biogenesis through a direct interaction with a nonribosomal factor in animals. A possible link between ribosome biogenesis and a transmembrane signal receptor in plants has yet to be discovered.
Figure 1.
cDNA microarray analysis of ribosomal proteins expression following fertilization in S. chacoense ovules. (A) Gene expression changes observed using nonparametric Anova testing (Kruskal- Wallis t-test), along with a Benjamini and Hochberg multiple testing correction algorithm of all the ribosomal protein genes available in our 7K microarray. (B) Cluster analysis of the differentially expressed ESTs corresponding to the ribosomal proteins in Table 1. The clones were classified based on the similarity of their expression profiles using a Self organization map (SOM) clustering. Three distinct SOM profiles (Group 1 to 3) can be distinguished. RNA extraction, probe preparation, cDNA array hybridization and data analysis were performed as described previously.27
and
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3724
References
- 1.van Nocker S, Ludwig P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics. 2003;4:50. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Germain H, Rudd S, Zotti C, Caron S, O'Brien M, Chantha SC, Lagace M, Major F, Matton DP. A 6374 unigene set corresponding to low abundance transcripts expressed following fertilization in Solanum chacoense Bitt, and characterization of 30 receptor-like kinases. Plant Mol Biol. 2005;59:515–532. doi: 10.1007/s11103-005-0536-8. [DOI] [PubMed] [Google Scholar]
- 3.Hennig L, Gruissem W, Grossniklaus U, Kohler C. Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol. 2004;135:1765–1775. doi: 10.1104/pp.104.043182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T. The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.) Plant J. 2005;41:660–672. doi: 10.1111/j.1365-313X.2005.02332.x. [DOI] [PubMed] [Google Scholar]
- 5.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 6.Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001;127:398–415. [PMC free article] [PubMed] [Google Scholar]
- 7.Tschochner H, Hurt E. Preribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 8.Baerson SR, Kaufman LS. Increased rRNA gene activity during a specific window of early pea leaf development. Mol Cell Biol. 1990;10:842–845. doi: 10.1128/mcb.10.2.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Kim SR, Chung YY, Lee JM, An G. Developmental and environmental regulation of two ribosomal protein genes in tobacco. Plant Mol Biol. 1994;25:761–770. doi: 10.1007/BF00028872. [DOI] [PubMed] [Google Scholar]
- 10.Larkin JC, Hunsperger JP, Culley D, Rubenstein I, Silflow CD. The organization and expression of a maize ribosomal protein gene family. Genes Dev. 1989;3:500–509. doi: 10.1101/gad.3.4.500. [DOI] [PubMed] [Google Scholar]
- 11.Stafstrom JP, Sussex IM. Expression of a ribosomal protein gene in axillary buds of pea seedlings. Plant Physiol. 1992;100:1494–1502. doi: 10.1104/pp.100.3.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams ME, Sussex IM. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- 13.Dresselhaus T, Cordts S, Heuer S, Sauter M, Lorz H, Kranz E. Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet. 1999;261:416–427. doi: 10.1007/s004380050983. [DOI] [PubMed] [Google Scholar]
- 14.Hulm JL, McIntosh KB, Bonham-Smith PC. Variation in transcript abundance among the four members of the Arabidopsis thaliana RIBOSOMAL PROTEIN S15a gene family. Plant Sci. 2005;169:267–278. [Google Scholar]
- 15.Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- 16.Popescu SC, Tumer NE. Silencing of ribosomal protein L3 genes in N. tabacum reveals coordinate expression and significant alterations in plant growth, development and ribosome biogenesis. Plant J. 2004;39:29–44. doi: 10.1111/j.1365-313X.2004.02109.x. [DOI] [PubMed] [Google Scholar]
- 17.Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 1994;13:3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 1999;18:490–499. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 20.Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell. 2005;17:2340–2354. doi: 10.1105/tpc.105.033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chantha SC, Matton DP. Underexpression of the plant NOTCHLESS gene, encoding a WD repeat protein, causes pleitropic phenotype during plant development. Planta. 2007 doi: 10.1007/s00425-006-0420-z. In press. [DOI] [PubMed] [Google Scholar]
- 22.de la Cruz J, Sanz-Martinez E, Remacha M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 2005;33:5728–5739. doi: 10.1093/nar/gki887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chantha SC, Emerald BS, Matton DP. Characterization of the plant Notchless homolog, a WD repeat protein involved in seed development. Plant Mol Biol. 2006;62:897–912. doi: 10.1007/s11103-006-9064-4. [DOI] [PubMed] [Google Scholar]
- 24.Cormier S, Le Bras S, Souilhol C, Vandormael-Pournin S, Durand B, Babinet C, Baldacci P, Cohen-Tannoudji M. The murine ortholog of notchless, a direct regulator of the notch pathway in Drosophila melanogaster, is essential for survival of inner cell mass cells. Mol Cell Biol. 2006;26:3541–3549. doi: 10.1128/MCB.26.9.3541-3549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royet J, Bouwmeester T, Cohen SM. Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 1998;17:7351–7360. doi: 10.1093/emboj/17.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigge PA, Weigel D. Arabidopsis genome: Life without notch. Curr Biol. 2001;11:R112–R114. doi: 10.1016/s0960-9822(01)00043-4. [DOI] [PubMed] [Google Scholar]
- 27.Gray-Mitsumune M, O'Brien M, Bertrand C, Tebbji F, Nantel A, Matton DP. Loss of ovule identity induced by overexpression of the fertilization-related kinase 2 (ScFRK2), a MAPKKK from Solanum chacoense. J Exp Bot. 2006;57:4171–4187. doi: 10.1093/jxb/erl194. [DOI] [PubMed] [Google Scholar]