Abstract
Regulation of genes by repression of transcription represents a virtually universal mechanism that underlies such diverse biological processes as restriction of expression of neuronal genes to neurons in mammals, and control of flowering in plants. However, while the molecular mechanisms of transcriptional gene silencing in animal systems are being intensively studied, our understanding of these processes in plants is very sparse and, because plants often utilize unique strategies to establish and maintain chromatin states, only limited use can be made of information available on epigenetic modifications in nonplant systems.
Key Words: Arabidopsis, chromatin remodeling, histone modification, gene repression, FLC
Our recent work identified two Arabidopsis factors, a SWIRM domain polyamine oxidase (PAO)-like protein, AtSWP1, and a plant-specific C2H2 zinc finger-SET domain histone methyltransferase (HMT), AtCZS, that interact with each other in plant cells and repress target gene(s) by histone hypoacetylation and generation of heterochromatic histone methylation marks. Thus, AtSWP1 and AtCZS were suggested to represent two main components of a corepressor complex. Here, we discuss these finding in the context of our knowledge of animal corepressor complexes, speculate on potential components of the AtSWP1/AtCZS corepressor complex that are unique to plants, and propose a working structure/function model of Arabidopsis PAO/HMT-containing corepressor complexes.
In eukaryotic cells, core histones undergo a variety of covalent modifications, which determine the structure and activity of the corresponding chromosomal region.1 These modifications promote conversion of the local chromatin into euchromatic or heterochromatic state, thus enhancing or restricting transcriptional activity, respectively, and are generated by chromatin-modifying complexes, which incorporate numerous enzymatic activities required for the specific modifications.2,3 Comprised of a large number of proteins and executing either transcriptional activation or repression tasks,2 these complexes play a crucial role during animal and plant growth and development.4,5 Yet, while this aspect of the eukaryotic cellular biology has been extensively investigated in animals, the studies of plant chromatin-modifying complexes are just beginning. Joining these research efforts, we have identified a novel type of transcriptional repressor complex which may act as one of the global regulators of plant gene expression and which combines both general, i.e., conserved between plants and animals, and plant-specific corepressor activities and components.6
Mammalian Polyamine Oxidase (PAO)/Lysine Demethylase 1 (LSD1)-Containing Corepressor Complex
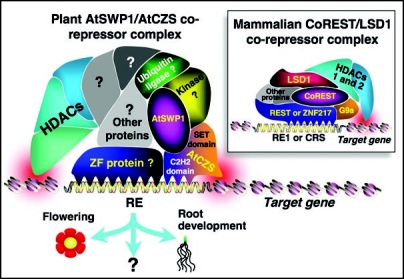
One of the well-studied examples of transcriptional repressors in animals is a multi-protein corepressor complex that specifically silences neuronal genes in nonneuronal cells (Fig. 1, inset). During mammalian development, a variety of neuronal genes, such as ion channels and neurotransmitter receptors, are silenced in nonneuronal tissues to allow correct cellular differentiation. A large group of these repressed genes, of which probably the best-known example is the type II voltage-dependant sodium channel,7,8 contain within their DNA sequence a 23 bp-long regulatory element, designated repressor element-1 (RE1).9,10 RE1 is directly recognized and bound by RE1-silencing transcription factor (REST),11 the DNA binding domain of which is represented by N-terminal C2H2-type zinc fingers motifs.8,11 Notably, REST does not possess any enzymatic activity and serves as a gene-targeting factor for the corepressor complex, while other components of the complex provide the actual enzymatic modules that covalently modify histones and repress expression of the target neuronal genes. These enzymatic activities include (1) histone deacetylases (HDACs) 1 and 2 which are thought to exist in complex12 with a REST-interacting protein CoREST;13 (2) LSD1, a protein containing two hallmark domains, SWIRM [Swi3p, Rsc8p, Moira14] that may anchor the tail of the histone H3 molecule,15 and PAO (polyamine oxidase)-like that acts as a histone H3 lysine demethylase;16 and (3) G9a, a SET [Su(var)3–9, Enhancer-of-zeste, Trithorax17]-domain protein with histone methyltransferase (HMT) activity.18,19 Besides these major components, other proteins, e.g., REST-associated mSin3, methyl-DNA binding factor MeCP2, etc.,20,21 copurify with the PAO/LSD1-containing corepressor complexes (Fig. 1, inset).
Figure 1.
Main components of the mammalian versus plant PAO-containing corepressor complexes. See the text for details.
Because the REST protein recognizes a unique RE1 sequence within the target gene(s), it makes biological sense that, instead of REST, other DNA binding zinc finger proteins may associate with the mammalian LSD1-containing corepressor complexes to direct them to gene groups with different response elements. Indeed, a zinc finger protein ZNF217 was shown to associate with CoREST/LSD1 complexes12,22 and bind a distinct 15 bp-long consensus recognition sequence (CRS) within the human E-cadherin promoter22 (Fig. 1, inset).
Plant PAO-Containing Co-Repressor Complex and Plant-Specific Histone Methyltransferase
Despite the importance of histone modifications in determination of the chromatin state and, therefore, cell fate during plant development,23,24 our knowledge about plant corepressor complexes in general, and PAO-containing corepressor complexes in particular, is virtually nonexistent. Only relatively recently an existence of a gene repression mechanism involving PAO-like protein has been provided by an observation that a plant homolog of the LSD1 gene, termed FLOWERING LOCUS D (FLD), represses, via histone hypoacetylation, FLOWERING LOCUS C (FLC), a negative regulator of flowering;25 the histone modifying machinery involved in the FLD-mediated repression of FLC, however, remains unknown. Unlike the composition of the protein complexes that modify plant histones, the histone modifications that index active or inactive chromatin are relatively well characterized. Specifically, Arabidopsis chromatin contains H3K4/H3K36 and H3K9/H3K27 methylation marks indexing active and inactive chromatin, respectively.26–31 Also, histone acetylation in plants has been characterized,32,33 and the family of plant HDAC proteins includes, based on sequence analysis of the Arabidopsis genome, three major groups: PD3/HDA1 and SIR2 conserved in eukaryotes and a plant-specific HD2 family.34
Recently, we have isolated two main components of a novel chromatin-modifying corepressor complex from Arabidopsis thaliana:—a SWIRM-domain PAO-like (SWP) protein AtSWP1, and a plant-specific C2H2 zinc finger PreSET-SET-PostSET-domain (CZS) protein AtCZS that likely acts as HMT (Fig. 1)—and demonstrated that they directly interact with each other in-planta (Fig. 2) and are required for generation of the H3K9 and H3K27 methylation marks on heterochromatin.6 AtCZS contains SET domain diagnostic of HMTs;35 thus, it presumably represents the actual enzyme that methylates H3K9 and H3K27 whereas the effect of AtSWP1 on these histone methylation marks is indirect, most likely through recruiting AtCZS. Because genetic inactivation of AtSWP1 and/or AtCZS alters not only histone methylation, but also acetylation,6 the AtSWP1/AtCZS corepressor complex most likely also contains as yet unidentified HDACs (Fig. 1).
Figure 2.
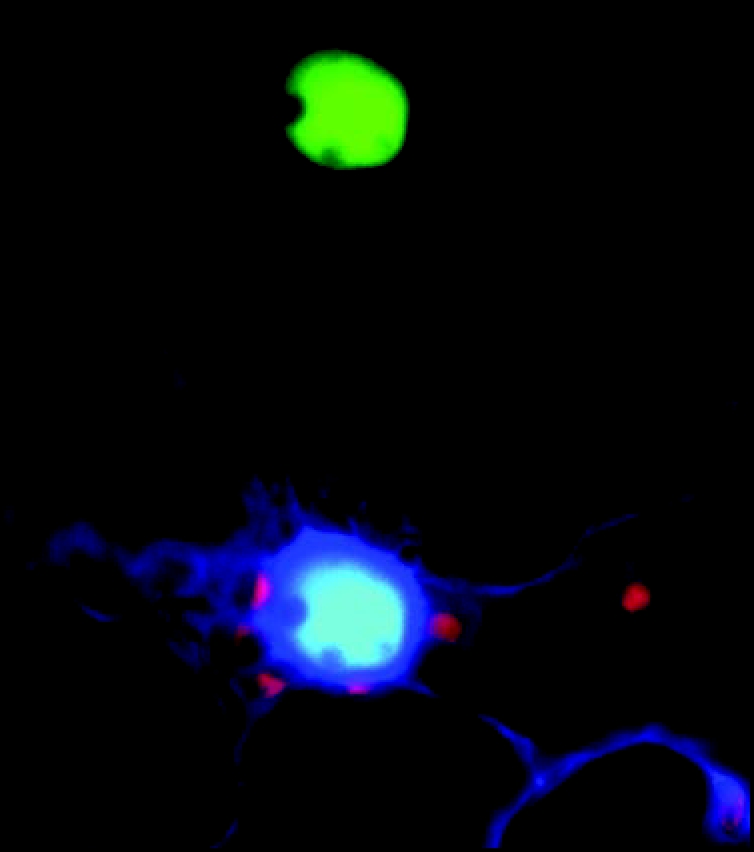
In planta interaction between AtSWP1 and AtCZS as detected by bimolecular fluorescence complementation (BiFC). (A) Reconstituted fluorescence of N-terminal and C-terminal halves of YFP fused to the interacting AtSWP1 and AtCZS proteins, respectively. (B) Merged images of the reconstituted YFP signal, co-expressed free CFP that identifies the cell nucleus and outlines the expressing cell, and plastid auto-fluorescence YFP signal is in green, CFP signal is in dark blue, overlapping YFP and CFP signals are in light blue, and plastid autofluorescence is in red. Images are a single confocal section. For further detail, see reference 6.
Interestingly, AtCZS represents a unique aspect of plant core- pressor complexes because of the combination of the SET and C2H2 zinc finger domains within the same protein molecule. Although conserved between evolutionarily-distant plant species such as monocots and dicots,6 it is found only in plants.35 In mammalian PAO-containing complexes,36 the AtCZS functionality is most closely represented by one of their zinc finger proteins, REST11,37 or ZNF217,12 and the PreSET-SET-PostSET HMT G9a protein that methylates H3K9 and/or H3K27.18,19 Thus, AtCZS likely combines the functionalities of the two different animal corepressor complex components in a single molecule (Fig. 1). In this scenario, AtCZS, similarly to REST or ZNF217, may play a role in initial association of the corepressor complex with regulatory elements of its target genes in the plant genome. Alternatively, the C2H2 zinc finger domain of AtCZS may simply augment and stabilize the interaction between the AtSWP1/AtCZS corepressor complex and its target gene whereas the binding specificity may be provided by another, as yet unidentified, DNA binding protein.
Another potential difference between the plant and animal PAO-containing corepressor complexes may lie in the functionality of the PAO-like protein itself. While both animal LSD1 and plant AtSWP1 exhibit a common conserved domain structure, i.e., N-proximal SWIRM domain and C-proximal PAO-like domain,6,16 only LSD1 possesses a detectable demethylase activity when tested in vitro using the purified recombinant proteins.6,16
A Working Model of the AtSWP1/AtCZS Corepressor Complex
Our working model of the AtSWP1/AtCZS corepressor is based on the notion that it may combine both general and plant-specific activities and components. For example, the presence of a PAO-like protein, SET-domain HMT, zinc finger proteins, and HDACs would represent a general feature, plant and animal. On the other hand, the specific PAO-like protein AtSWP1 may lack the demethylase activity, and the AtCZS is a plant-specific HMT which is not found in animal complexes. It is tempting to speculate that another potential plant-specific feature of the AtSWP1/AtCZS complex may be association with plant-specific HD2 HDACs. Moreover, plant corepressors may contain multiple species of the AtSWP protein family, which comprises four members in Arabidopsis,16 and one of these proteins may be a functional histone demethylase. Finally, plant complexes may contain novel functions, such as kinases and ubiquitin ligases (Fig. 1), which have been identified to bind AtSWP1 in our ongoing yeast two-hybrid protein interaction screens (unpublished data). Because histones are known to undergo phosphorylation38 and ubiquitination,39 it is possible that the AtSWP1/AtCZS corepressor complex may be able to affect an exceptionally wide spectrum of histone modification, from methylation/demethylation to deacetylation, to ubiquitination and to phosphorylation, making it a truly multifunctional chromatin modifier.
Another important and still unresolved aspect of the AtSWP1/AtCZS complex is the range of its target genes. To date, we have documented one AtSWP1/AtCZS target gene, the FLC negative regulator of flowering, and the corresponding flower timing delay phenotypes of the Arabidopsis null mutants in AtSWP1 and AtCZS.6 However, these mutants exhibited other, unrelated to FLC, phenotypic changes, such as altered root length, indicating existence of additional target genes of the AtSWP1/AtCZS complex (Fig. 1). Indeed, our microarray analyses of both mutant lines identified over 70 genes the expression of which was upregulated 4–8 fold in the mutants as compared to the parental, wild-type plants (unpublished data). A significant subset of these upregulated genes, including those predicted to participate in root development, was shared between the two mutants whereas no genes were detected that showed significant down-regulation in either of the mutants. These observations are consistent with the role of AtSWP1 and AtCZS as gene repressors acting in concert in the same multiprotein complex. Furthermore, they suggest a role for the AtSWP1/AtCZS corepressor complex in global regulation of plant gene expression.
Acknowledgements
The work our laboratory is supported by grants from NIH NSF, USDA, BARD, BSF and CDR-USAID to V.C.
note
Figure 2 can be seen in color at: www.landesbioscience.com/journals/psb/abstract.php?id=3726
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/abstract.php?id=3726
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 5.Goodrich J, Tweedie S. Remembrance of things past: Chromatin remodeling in plant development. Annu Rev Cell Dev Biol. 2002;18:707–746. doi: 10.1146/annurev.cellbio.18.040202.114836. [DOI] [PubMed] [Google Scholar]
- 6.Krichevsky A, Gutgarts H, Kozlovsky SV, Tzfira T, Sutton A, Sternglanz R, Mandel G, Citovsky V. AtCZS, an Arabidopsis C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Dev Biol. 2007:301. doi: 10.1016/j.ydbio.2006.11.012. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: A model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 8.Tapia-Ramirez J, Eggen BJ, Peral-Rubio MJ, Toledo-Aral JJ, Mandel G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc Natl Acad Sci USA. 1997;94:1177–1182. doi: 10.1073/pnas.94.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maue RA, Kraner D, Goodman RH, Mandel G. Neuron-specific expression of the rat brain type II sodium channel gene is directed by upstream regulatory elements. Neuron. 1990;4:231–233. doi: 10.1016/0896-6273(90)90097-y. [DOI] [PubMed] [Google Scholar]
- 10.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 11.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 12.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L, Iyer LM. The SWIRM domain: A conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 2002;3:0039.1–0039.7. doi: 10.1186/gb-2002-3-8-research0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 19.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 20.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. The corepressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 22.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: Identification of a ZNF217 consensus recognition sequence. Oncogene. 2006 doi: 10.1038/sj.onc.1210126. In press. [DOI] [PubMed] [Google Scholar]
- 23.Loidl P. A plant dialect of the histone language. Trends Plant Sci. 2004;9:84–90. doi: 10.1016/j.tplants.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Wagner D. Chromatin regulation of plant development. Curr Opin Plant Biol. 2003;6:20–28. doi: 10.1016/s1369526602000079. [DOI] [PubMed] [Google Scholar]
- 25.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 26.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I. Histone modifications in Arabidopsis—High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003;33:471–480. doi: 10.1046/j.1365-313x.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- 28.Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 30.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol. 2005;7:1156–1160. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- 32.Verbsky ML, Richards EJ. Chromatin remodeling in plants. Curr Opin Plant Biol. 2001;4:494–500. doi: 10.1016/s1369-5266(00)00206-5. [DOI] [PubMed] [Google Scholar]
- 33.Li G, Hall TC, Holmes-Davis R. Plant chromatin: Development and gene control. Bioessays. 2002;24:234–243. doi: 10.1002/bies.10055. [DOI] [PubMed] [Google Scholar]
- 34.Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucl Acid Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of corepressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 37.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 39.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 40.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]