Abstract
We previously screened genes that were transcriptionally activated during the early stage of wound response in tobacco plants (Nicotiana tabacum), and isolated a particular clone, which encoded a membrane-located protein, designated as NtC7. Upon overexpression in tobacco plants, NtC7 conferred a marked tolerance to osmotic stress, suggesting it to be involved in maintenance of osmotic adjustments. In this study, we searched for proteins which interact with NtC7 by the yeast two-hybrid screening, and isolated a clone encoding phosphoinositide-specific phospholipase C, designated as NtPI-PLC. Physical interaction between NtC7 and C2 domain of NtPI-PLC was confirmed by the pull-down assay. Expression of fused protein to green-fluorescence protein in onion epidermal cell layers indicated both proteins to predominantly localize to the plasma membrane. Their interaction in planta was shown by the bimolecular fluorescence complementation, which exhibited a clear fluorescence of reconstituted yellow fluorescence protein. Transcripts of NtC7 and NtPI-PLC were markedly increased 30 to 60 min after wounding. PI-PLC is one of key enzymes in metabolism of inositol phospholipids, which function in signal transduction and also in response to stresses including osmotic changes. It was shown to localize to plasma-membrane and, to a lesser extent, to cytosol. However, molecular mechanism of membrane localization has remained to be determined, because of the apparent lack of domains for membrane association. The present results suggest that one of such mechanisms is tethering NtPI-PLC to the plasma membrane through interaction with NtC7, which possesses a transmembrane domain at the C-terminus.
Key words: Nicotiana tabacum, osmotic stress, phosphoinositide-specific phospholipase C, PI-PLC-linker protein, wounding
Wounding is one of the severest stresses for plants, causing not only direct mechanical injury to plant body but also secondary physiological effects including metabolic disturbance and susceptibility to pathogen infection. Plants have developed various systems to cope with these detrimental effects of wounding by expressing a set of stress-responsive genes. In our previous studies, we screened by the differential display method for tobacco genes, which were expressed at the early stage of wounding, and initially identified 28 clones, some of which were further characterized.1,2 One clone, designated as NtC7, was found to encode a membrane-located protein with an apparent molecular mass of 34 kDa (accession no. AB087235).2 Its transcripts were undetectable under non-stressed condition, but heavily accumulated upon wounding and osmotic stress. Transgenic tobacco plants overexpressing NtC7 exhibited a clear tolerance to a high concentration of mannitol, but not to sodium chloride. It was suggested that NtC7 was involved in osmotic adjustment system independently of ion homeostasis.2
Phosphlipases catalyze hydrolysis of phospholipids, which constitute the backbone of biomembrane, and serve as the source of signaling molecules.3–5 Phosphoinositide-specific phospholipase C (PI-PLC) hydrolyzes phosphatydilinositols releasing inositol triphosphates, which are one of the key components in signal transduction pathways.6 In plants, PI-PLC has been shown to be involved in response to osmotic stress, abscisic aid, light, gravity and pathogen attack, suggesting its fundamental cellular function.5–7 Immuno-staining and biochemical fractionation assays indicated it to be predominantly localized to membrane, and also to cytoplasmic fraction.8,9 Structurally, plant PI-PLC resembles animal proteins except that it lacks the pleckstrin homology (PH) domain, which is commonly found in animal PLCs and required for interaction with the plasma membrane.10 It has thus remained to be determined how plant PI-PLC could be associated with plasma membrane. In this addendum, we describe that, in tobacco plants, PI-PLC specifically interacts with the membrane resided NtC7, and therefore that the latter may serve as a linker to tether PI-PLC to the plasma membrane. This could be a novel mechanism for membrane association of PI-PLC that possesses no apparent domains to directly interact with membrane structure.
Tobacco plants (Nicotiana tabacum cv. Xanthi nc) were grown on soil in a growth cabinet and wounding was given to healthy leaves by cutting.2 Yeast two-hybrid assay was performed using commercially available system (Matchmaker two-hybrid system 2; Clonetech). A cDNA library was constructed from transcripts isolated from wounded or unwounded tobacco leaves using pGADT7 (Clonetech).11 Using the full-length NtC7 bait plasmid, approximately 8.4 × 108 clones were screened on Leu/Trp/His/Ala deficient SD medium, and β-galactosidase assay was performed as described.11 Since the initially isolated cDNA was a fragment of 396 bp, a full length cDNA was prepared by PCR using appropriate sets of primers designed after the sequence from N. rustica PI-PLC cDNA (accession no. X95877). Pull-down binding assay was performed with GST-tagged NtC7 and His-tagged C2 domain (amino acid positions 466–587) of NtPI-PLC. Subcellular localization was eaxamined using full-length cDNAs of NtC7 and NtPI-PLC cloned into the CaMV35S-sGFP(S65T)-nos vector.12 Gold particles coated with resulting plasmids were bombarded into onion epidermal cells and GFP fluorescence was imaged by a confocal laser-scanning microscope LSM510 (Carl Zeiss). Bimolecular fluorescence complementation (BiFC) analysis using yellow fluorescence protein (YFP) system was performed as described.13,14 The full-length NtC7 cDNA was subcloned into pSY736 containing the N-terminus of YFP (YN), or pSY735 containing the C-terminus of YFP (YC) vectors, and the full-length NtPI-PLC cDNA was subcloned into pSY728 (YN) or pSY738 (YC) vectors.15 Vectors without inserts were used under the same combination as the control. Resulting plasmids were bombarded into onion epidermal cell layers under appropriate combinations, and fluorescence from YFP was imaged by a fluorescence microscope (AX70, Olympus).
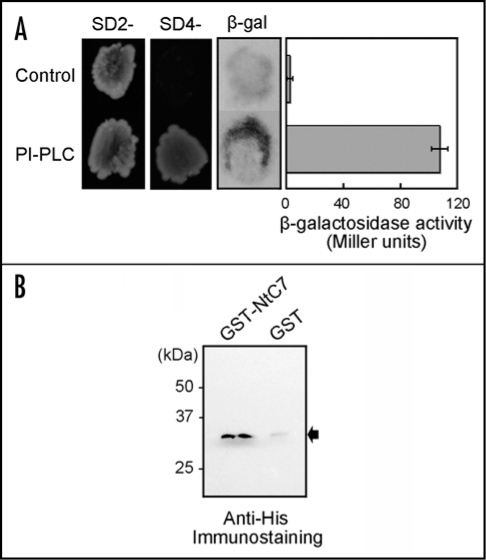
Since membrane-located proteins are considered to often function through protein-protein interactions, we attempted to identify proteins directly interacting with NtC7 by the yeast two-hybrid screening. Among 80 clones which were initially positive in both histidine and β-galactosidase complementation assays, two were found to encode a polypeptide resembling the C2 domain of PI-PLC from Nicotinia rustica. Since the identified cDNA clone was a 5′-truncated product, the full length cDNA was isolated from the tobacco cDNA library. The cDNA was 2083 bp in length, and encoded a polypeptide with 588 amino acids, showing a 94% similarity to N. rustica PI-PLC (accession no. X95877). The protein was consequently designated as NtPI-PLC under the accession number EF520286. The initially isolated clone was 396 bp encoding a polypeptide with 132 amino acids, which corresponded to the amino acid positions between 435 and 567 of NtPI-PLC. As the C2 domain of NtPI-PLC is a polypeptide with 128 amino acids between positions 460 and 588 at the C-terminus, the isolated clone covered the majority of the C2 domain. To confirm the specificity of the interaction, a plasmid containing the C2 domain (amino acid positions, 466–587) was constructed and co-transformed with either GBD-NtC7 (bait plasmid) or the pBD-GAL4 Cam vector into yeast Y190. Only the transformants containing plasmids encoding both C2 domain and NtC7 were positive in the histidine and β-galactosidase reporter assays (Fig. 1A), indicating the interaction between them to indeed be specific. The observed interaction between NtC7 and C2 domain of NtPI-PLC in the yeast two-hybrid system was confirmed by an in vitro pull-down assay (Fig. 1B). His-tagged C2 domain of NtPI-PLC was applied to a glutathione-Sepharose column containing NtC7-GST or GST proteins, eluted with a buffer containing reduced glutathione, separated by SDS-polyacrylamide gel electrophoresis and subjected to immuno-blot assays with anti-His-tag antibodies. The results clearly showed that signal was only detectable upon incubation with NtC7-GST but not with GST protein alone, indicating specific binding of the C2 domain of NtPI-PLC to NtC7 (Fig. 1B).
Figure 1.
Idintification of NtC7 interacting protein. (A) Yeast two-hybrid assay. Yeast Y190 cells were co-transformed with GBD-NtC7 and GAD-NtPI-PLC fragment. The transformant was planted on SD agar supplemented with an amino acid mixture depleted of tryptophan and leucine (SD/2-), or on SD agar supplemented with an amino acid mixture depleted of tryptophan, leucine, alanine and histidine (SD/4-), and assayed for β-galactosidase by the filter lift method (β-gal) (indicated as PI-PLC on the left). The β-galactosidase activity was estimated using O-nitriphenyl-β-d-galactopyranoside as the substrate and expressed in Miller units. As the negative control, BD-pGBKT7-53 was co-transformed with AD-pGADT7-Rec (indicated as Control on the left). Values are from triplicates with standard deviation. (B) Pull-down assay. Interaction was tested using GST-fused full length NtC7 and His-tagged C2 domain of NtPI-PLC proteins. NtC7-GST proteins were bound to a glutathione-column and His-tagged C2 domain was added. After elution with reduced glutathione, proteins were separated on SDS-polyacrylamide gels and the C2 domain was detected by anti-His-tag antibodies (indicated by an arrow). Note that, although a relative molecular mass of the C2 polypeptide (121 amino acids) was approximately 16 kD, a fusion protein with His-tag often migrates slow on polyacrylamide gel electrophoresis due to the strong basic charge of histidine residues, showing apparently higher molecular mass with extra 10 to 20 kD.
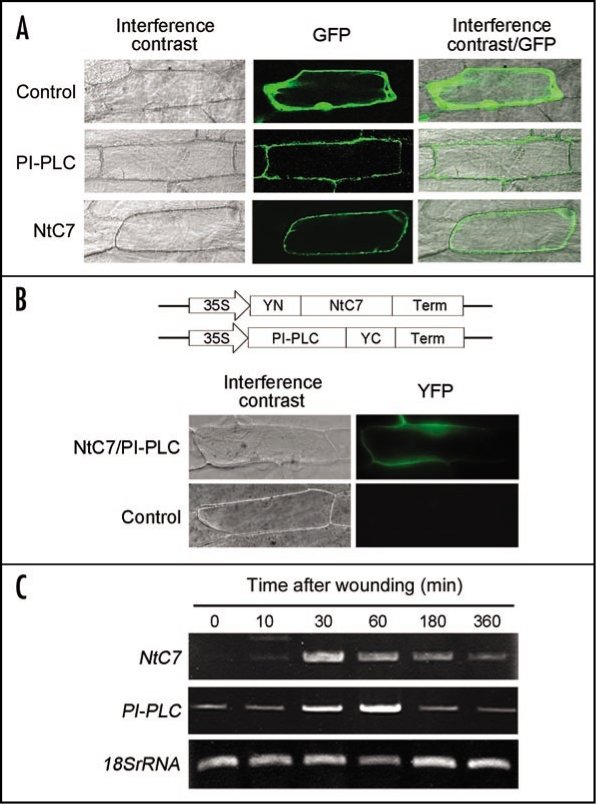
Intracellular localization of NtC7 and NtPI-PLC was first examined using green fluorescence protein (GFP)-fusion proteins, driven by the cauliflower mosaic virus (CaMV) 35S promoter. Transient expression by particle bombardment into onion epidermal cell layers showed fluorescence from the control 35S-GFP to be observed throughout cells (Fig. 2A). Fluorescence from both NtC7-GFP and NtPI-PLC-GFP was clearly observed in membrane fraction (Fig. 2A), suggesting that these proteins reside and possibly interact each other at plasma membrane. Interactions between NtC7 and NtPI-PLC in planta were directly examined by the bimolecular fluorescence complementation (BiFC) analysis, in which active yellow fluorescence protein (YFP) is reconstituted only when non-fluorescent N-terminal (YN) and C-terminal (YC) YFP fragments are brought together by protein-protein interactions.15,16 Experimentally, either NtC7 or NtPI-PLC was fused to either YN or YC at either its C- or N-terminus, so that all theoretical combinations among proteins could be examined.14 Among eight such possible combinations, we initially constructed a pair of plasmids, in which NtC7 was fused to the C-terminus of the YN fragment (YN-NtC7) and NtPI-PLC to the N-terminus of the YC fragment (NtPI-PLC-YC) (Fig. 2B, upper). When they were bombarded into onion epidermal cells, active YFP was apparently reconstituted, showing clear fluorescence (Fig. 2B, lower). Since the control combination without NtC7 and NtPI-PLC inserts did not yield fluorescence, it was clear that observed YFP fluorescence was the result of specific interaction between the two proteins. These two observations using GFP and BiFC (Fig. 2A and B) hence strongly suggested that NtC7 and NtPI-PLC interact in planta at plasma membrane.
Figure 2.
Properties of NtPI-PLC and NtC7. (A) Intracellular localization. Onion epidermal cell layers were bombarded with gold particles coated with GFP alone (Control), NtC7-GFP (NtC7) or NtPI-PLC-GFP (PI-PLC) and observed for epifluorescence of GFP (GFP) under a confocal laser-scanning microscope. Fluorescence and bright field images (interference contrast) were merged (interference contrast/GFP). (B) Bimolecular fluorescence complementation assay. Indicated BiFC vectors (upper) were introduced into onion epidermal cell by the particle bombardment method, and observed under a fluorescence microscope equipped with the YFP fluorescent module (lower, NtC7/PI-PLC). Empty vectors containing no inserts were used as the control (lower, Control). YFP (YFP) shows BiFC results and bright field images are also shown (interference contrast). Note that slight difference of images between A and B is due to the different microscope used. (C) Expression profile. Induction of NtC7 and NtPI-PLC transcripts by wounding was examined by RT-PCR using RNAs isolated from wounded leaves at indicated time period. Two µg of total RNA was first reverse-transcribed with oligo(dT)15, then amplified with specific primers designed for NtC7, NtPI-PLC or 18S ribosomal RNA, and separated by agarose gel electrophoresis.
Expression profile of NtC7 and NtPI-PLC was examined by semi-quantitative RT-PCT. Healthy tobacco leaves were wounded by cutting and total RNA was extracted at appropriate time periods. Transcripts of NtC7 were not detected under non-stressed condition, but began to heavily accumulate 30 min after injury, and gradually declined by 6 h (Fig. 2B). In contract, transcripts of NtPI-PLC were constitutively accumulated at a low level under non-stressed condition. However, their level distinctly increased 30 min after wounding, reached maximal level at 1 h, and declined thereafter to the basal level by 6 h (Fig. 2B). These results suggested that both NtC7 and NtPI-PLC proteins were coordinately synthesized upon wounding stress, although NtPI-PLC appeared to be constantly produced at a low level regardless the stress.
The NtC7 consists of 308 amino acids with a relative molecular mass of 34 kD.2 The N-terminus region with 147 amino acids resembles the receptor domain of receptor-like kinases (RLK), and the C-terminus region contains a transmembrane domain with a stretch of 17 amino acids (positions 275–291). Membrane localization was confirmed by transient assays using GFP-fusion protein in onion epidermis cells. Transgenic tobacco plants overexpressing NtC7 showed an elevated tolerance against osmotic stress.2 Since our finding of NtC7 in 2003, similar proteins have been identified from several plant species, including potato (accession nos. CV497517, CV507089), tomato (accession no. AI775018) and pepper (accession no. CA516173). In addition to osmotic stress, NtC7 was shown to be transcriptionally activated during hypersensitive response.17 Based on these observation, we speculated that NtC7 and NtC7-like proteins might function in sensing and/or responding to osmotic changes at plasma membrane. If this were the case, identification of interacting protein(s) with NtC7 was conceivably critical to further understand the molecular mechanism of osmostress response in plants.
Subsequent screening revealed that PI-PLC was one of such protein(s). PI-PLC is a critical enzyme involved in inositol catabolism in both animal and plant cells.5,6 In animals, its best-known function is the hydrolysis of phosphatidylinositol(4,5) bisphosphate to produce inositol(1,4,5)triphosphate and diacylglycerol, which are the major cellular messengers. In plants, less information is available, but its involvement in calcium mobilization and oscillation appears to be important.6 Many physiological features related to calcium metabolism have been reported to be linked to PI-PLC, as exemplified by abscisic acid18,19 and cytokinine7 responses, hyperosmotic stress20,21 and developmental process.3 Plant PI-PLC contains four conserved regions, EF-hand, X and Y, and C2 domains, these being essentially identical with animal proteins.4 A notable difference is that plant enzyme does not possess a pleckstrin homology (PH) domain, which is found in most animal proteins, and indispensable for both membrane association and displacement.10
The lack of the PH domain has raised a question as to how plant PI-PLC associates with plasma membrane.4 The present finding gives a clue, showing that the C2 domain of NtPI-PLC efficiently binds to a linker protein, NtC7, which possesses a transmembrane domain at its C-terminus. The C2 domain of animal PLC-δ1, which most resembles plant PI-PLCs, contains several Asp and Asn residues, that are important to recognize calcium ions and phosphatidylserines localized in plasma membrane.22,23 These observations suggest that the C2 domain of animal PI-PLCs directly associates to plasma membrane through amino acid interaction under calcium-dependent manner.4,22 In contrast, our finding points to that plant PI-PLC can be indirectly retained to plasma membrane through protein-protein interaction between the C2 domain and a membrane-resided linker protein. This idea has correctly been proposed by Mueller-Roeder and Pical in 2002, describing that the C2 domain of plant PI-PLCs may be sufficient for membrane-binding.
Overall, the present results suggest that one of the biological functions of NtC7 is to serve as an anchor to retain PI-PLC at plasma membrane. Although the possibility that NtC7 directly regulates PI-PLC activity through molecular interaction can not currently be excluded, the former idea accounts for our previous observation, showing overexpression of NtC7 in tobacco plants conferred a significant tolerance against osmotic stress.2 It is conceivable that high level of NtC7 tethered more PI-PLC to plasma membrane, thereby strengthening its function.8,9,24
Acknowledgements
The authors thank Dr. Yutaka Kodama (Kyushu University, Japan) for valuable discussion on BiFC experiments, and Ms. Yuka Yamamoto (Nara Institute of Science and Technology) for manuscript preparation. This work was partly supported by a grant from the Japan Society for the Promotion of Science.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7222
References
- 1.Hara K, Yagi M, Koizumi N, Kusano T, Sano H. Screening of wound-responsive genes identifies an immediate-early expressed gene encoding a highly charged protein in mechanically wounded tobacco plants. Plant Cell Physiol. 2000;41:684–691. doi: 10.1093/pcp/41.6.684. [DOI] [PubMed] [Google Scholar]
- 2.Tamura T, Hara K, Yamaguchi Y, Koizumi N, Sano H. Osmotic stress tolerance of transgenic tobacco expressing a gene encoding a membrane-located receptor-like protein from tobacco plants. Plant Physiol. 2003;131:454–462. doi: 10.1104/pp.102.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson JM, Perera IY, Heilmann I, Persson S, Boss W. Inositol signaling and plant growth. Trends Plant Sci. 2000;5:252–258. doi: 10.1016/s1360-1385(00)01652-6. [DOI] [PubMed] [Google Scholar]
- 4.Muller-Roeber B, Pical C. Inositol phospholipids metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipids kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 2002;130:22–46. doi: 10.1104/pp.004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 6.Wang X. Plant phospholipases. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:211–231. doi: 10.1146/annurev.arplant.52.1.211. [DOI] [PubMed] [Google Scholar]
- 7.Repp A, Mikami K, Mittmann F, Hartmann E. Phosphoinositide-specific phospholipase C is involved in cytokinin and gravity responses in the moss Physcomitrella patens. Plant J. 2004;40:250–259. doi: 10.1111/j.1365-313X.2004.02205.x. [DOI] [PubMed] [Google Scholar]
- 8.Shi J, Gonzales RA, Bhattacharyya MK. Characterization of a plasma membrane-associated phosphoinositide-specific phospholipase C from soybean. Plant J. 1995;8:381–390. doi: 10.1046/j.1365-313x.1995.08030381.x. [DOI] [PubMed] [Google Scholar]
- 9.Otterhag L, Sommarin M, Pical C. N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C acrivity in Arabidopsis thaliana. FEBS Lett. 2001;497:165–170. doi: 10.1016/s0014-5793(01)02453-x. [DOI] [PubMed] [Google Scholar]
- 10.Paterson HF, Savopoulos JW, Perisi O, Cheung R, Ellis MV, Williams RL. Phospholipase Cδ1 requires a pleckstrin homology domain for interaction with the plasma membrane. Biochem J. 1995;312:661–666. doi: 10.1042/bj3120661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MH, Sano H. Attenuation of hypersensitive response by an ATPase associated with various cellular activities (AAA) protein through suppression of a small GTPase, ADP ribosylation factor in tobacco plants. Plant J. 2007;51:127–139. doi: 10.1111/j.1365-313X.2007.03124.x. [DOI] [PubMed] [Google Scholar]
- 12.Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 13.Yano A, Kodama Y, Koike A, Shinya T, Kim HJ, Matsumoto M, Ogita S, Wada Y, Ohad N, Sano H. Interaction between methyl-CpG binding protein and Ran GTPase during cell division in tobacco cultured cells. Ann Bot. 2006;98:1179–1187. doi: 10.1093/aob/mcl211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodama Y, Shinnya T, Sano H. Dimerization of N-methyltransferases involved in caffeine biosynthesis. Biochimie. 2008;90:547–551. doi: 10.1016/j.biochi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 16.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 17.Ghannam A, Jacques A, De Ruffray P, Baillieul F. Identification of tobacco ESTs with a hypersensitive response (HR)-spesific pattern of expression and likely involved in the induction of the HR and/or localized acquired resistance (LAR) Plant Physiol Biochem. 2005;43:249–259. doi: 10.1016/j.plaphy.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Staxen I, Pical C, Montgomery LT, Gray JE, Hertherington AM, McAinsh MR. Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositidespacific phospholipase C. Proc Natl Acad Sci USA. 1999;96:1779–1784. doi: 10.1073/pnas.96.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt L, Mills LN, Pical C, Leckie CP, Aitken FL, Kopka J, Mueller-Roeber B, McAinsh M, Hetherington AM, Gray JE. Phospholipase C is required for the control of stomatal aperture by ABA. Plant J. 2003;34:47–55. doi: 10.1046/j.1365-313x.2003.01698.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1995;94:3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parre E, Ghars MA, Leprince AS, Thiery L, Lefebvre D, Bordenave M, Richard L, Mazars C, Abdelly C, Savoure A. Calcium signaling via phospholipase C is essential for proline accumulation upon ionic but not noninonic hyperosmotic stresses in Arabidopsis. Plant Physiol. 2007;144:503–512. doi: 10.1104/pp.106.095281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 23.Ananthanarayanan B, Das S, Rhee SG, Murray D, Cho W. Membrane targenting of C2 domains of phospholipase C-δ isoforms. J Biol Chem. 2001;277:3568–3573. doi: 10.1074/jbc.M109705200. [DOI] [PubMed] [Google Scholar]
- 24.Drobak BK, Watkins PAC. Inositol(1,4,5)triphosphate production in plant cells: an early response to salinity and hyperosmotic stress. FEBS Lett. 2000;481:240–244. doi: 10.1016/s0014-5793(00)01941-4. [DOI] [PubMed] [Google Scholar]