Abstract
Human BRCA1 (BRreast CAncer susceptible gene1) is known to involve in cell cycle control, transcriptional regulation, DNA recombination, DNA repair and many other processes. hBARD1 (BRCA1-Associated Ring Domain 1) forms heterodimer via its N-terminal conserved RING domain with BRCA1. In Arabidopsis, two genes, At4g21070 and At1g04020, that share N-terminal RING domain and C-terminal BRCT (for BRCA1 C-Terminal) domains with no substantial similarities for other motifs, have been identified. AtBRCA1 was induced by γ-ray while AtBARD1 was required for DNA repair. Recently, we find that AtBARD1 may function to confine WUS transcription in the shoot apical meristem organization center, together with the ATPase-dependent chromatin remodeling factor, SYD. In bard1–3 Arabidopsis knockout mutant, WUS was released to the outer layers and expressed at extremely high level comparing to wild-type. Our data suggest that BARD1 mainly function as a REPRESSOR OF WUSCHEL1 (ROW1). Extensive motif analyses carried out here showed that ROW1 possesses substantial sequence identity with a reported transcription repressor, MLL and also a potential PHD domain which recognizes histone tail codes, in its uncharacterized middle region. We suggest that ROW1 represses transcription in a chromatin-related mechanism.
Key words: Arabidopsis, BARD1, ROW1, shoot apical meristem, chromatin remodeling
hBRCA1 has been extensively studied for more than a decade and is an essential component for many cellular processes.1–4 Multiple functions of BRCA1 may be attributed to its specific protein structure. Its N-terminal RING domain contains a classical C3HC4 zinc-finger which is active in the ubquitination-related E3 ligase protein degradation pathway.5,6 Also, through its RING-mediated interaction, hBRCA1 is involved in cell cycle control by coupling with cyclins, cyclin-dependent kinases and the transcription factor E2F-4.7 Two tandem-repeated BRCT domains that are highly-conserved in many important components in DNA repair processes, such as XRCC, 53BP1 and MDC1,8 are located at its C-terminal. BRCTs are known to be involved in a phosphorylation-mediated transcription repression with the transcriptional co-repressor Ct-IP,9,10 in a chromatin remodeling mechanism together with DNA helicase BACH1,11,12 and also in SWI/SNF1 complexes.13 BARD1 contains a RING domain in its N-terminal and two tandem BRCT domains in its C-terminal, and was reported to form a heterodimer with BRCA1 via conserved RING domain.14 The BRCA1-BARD1 complex was suggested to play pivotal roles in cancer suppression.6 However, BARD1, independent of BRCA1, can also function as a cancer suppressor since inherited mutations of BARD1 was demonstrated to be involved in endometrial cancer, as well as control of cell proliferations of mammary glands.15
Plant homologs of BRCA1 and BARD1 have been characterized in Arabidopsis. In 2003, At4g21070 was named AtBRCA1 since it showed a 30% sequence identity with hBRCA1 at the P300/TBP interacting site (a 25 amino acid-long motif) located in the middle of the protein, in addition to the conserved N-terminal RING and C-terminal BRCT domains.16 A similar gene on Arabidopsis chromosome 1 (At1g04020) with almost identical BRCT and RING domain structures was named BARD1 because of its possible involvement in DNA repair, consistent with its function in mammals.4,17 In terms of first-order amino acid sequence identity, AtBRCA1 and AtBARD1 share no substantial homolog apart from these conserved motifs. In fact, AtBRCA1 lacks the SQ-cluster domain (SCD) that is typical of hBRCA1, and AtBARD1 lacks the Ankyrin (ANK) motif typical of hBARD1.15,17
In plants, all above-ground tissue develops from stem cells located in shoot apical meristem (SAM). The WUS-CLV3 feedback loop is responsible for the organization and maintenance of the stem cell poll.18–20 In the bard1–3 knock-out mutant, WUS was released to the outer layers and expressed 238-fold comparing to wild type.21 A specific WUS promoter region was recognized by nuclear protein extracts obtained from wild-type plants, and this protein-DNA complex was recognized by antibodies against BARD1. The double mutant (wus-1 bard1–3) showed prematurely terminated SAM structures identical to those of wus-1. Similar phenotype was observed in BARD1 overexpression lines since WUS transcripts were reduced significantly in these plants. These data suggest that BARD1 regulates SAM organization and maintenance by limiting WUS expression to the organizing center. Further analyses revealed that BARD1 might function together with SYD, the chromatin remodeling factor, which is specifically recruited to the WUS promoter.21–22
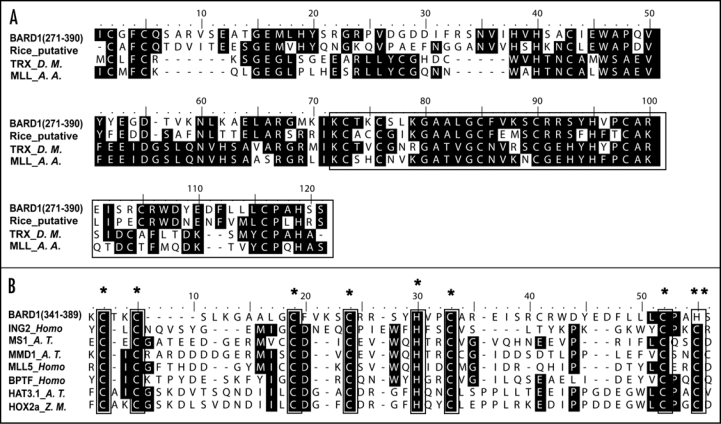
Here extensive motif scan carried out on the middle region of At1g04020 reveals more evidences for it to serve as a functional repressor (Fig. 1A). The region from residues 271 to 390 of At1g04020 showed about 33% sequence identity or 54% similarity with MLL in yellow fever mosquito (Aedes aegypti), and 38% identity or 53% similarity with TRX in drosophila. MLL and TRX are involved in chromatin modifications and in the interaction between chromatin and proteins. MLL, for mixed-lineage leukemia protein, possesses a transcription repression domain, and is associated with acute myeloid and lymphoid leukemia. This protein shares high sequence homology with DNA methyltransferase and interacts with histone deacetylase 1, indicating that it may function at the chromatin level.23,24 TRX was suggested to be a histone methyltransferase (HMTase) that methylates distinct lysine residues in the N-terminal tail of histone H3. Modification of histone may inhibit transcription activity either by recruiting repression complex or by blocking the access of transcription factors to the promoter region.25
Figure 1.
The Middle Region of ROW1 Shows Substantial Sequence Similarity with Proteins of Different Functions. (A) Multiple sequence alignment of ROW1 (Amino acid 271–390) with TRX (trithorax, gene ID: 41737),31 MLL (mixed-lineage leukemia protein, gene ID: 5563643)32 and a putative rice protein (Os 04g0512400). (B) ROW1 contains a variant form of the PHD domain. Sequences framed in the box in (A) are further aligned with several characterized PHD (Cysteine4-Histidine-Cysteine3) motifs. ING2, inhibitor of growth family member 2;33 MS1, MALE STERILITY1 in Arabidopsis;34 MMD1, MALE MEIOCYTE DEATH1 in Arabidopsis;35 HAT3.1, an Arabidopsis homeobox protein;36 MLL5, human MIXED-LINEAGE LEUKEMIA5 protein;37 HOX2a, a maize homeobox gene.38 *denotes identical amino acids; ** denotes similar amino acid.
The last motif of the aligned sequences, highlighted by a rectangle shown in Figure 1A, is a potential PHD (Plant Homeodomain) motif that was reported to be a specialized histone tail code reader. Proteins containing this domain participated in transcription regulation through specific interactions with epigenetic markers, trimethylated lysine 4 on the N-terminal tail of histone 3 (H3K4).26–29 When we align ROW1 with other well-known PHD motifs, we find that different from the classical C3H2C2 consensus pattern, the last C in ROW1 is replaced by H (Fig. 1B), which is a variant form of PHD that retains the ability to bind zinc atoms.30 When gene transcription is turned “on”, the nucleosome structures need to be unpacked, or changed into a “loose” status from a “tight” confirmation, for easy access of transcription factors to the promoter region. Many epigenetic markers on the histone are responsible for the maintenance of “loose” or “tight” state of the nucleosome structure, and also for determining the affinity of transcription factors to the promoter.25 Based on our current understanding, ROW1 may repress transcription of WUS by modulating chromatin structure, possibly together with SYD and through the PHD domain. We suggest that detailed exploration of WUS1 expression may lead to new insight into mechanisms leading towards the regulation of transcription factors at large.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7312
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7:937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- 3.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 4.Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr Opin Cell Biol. 2003;15:345–350. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 5.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 6.Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Shao N, Ding QM, Cui J, Reddy ES, Rao VN. BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases. Oncogene. 1997;15:143–157. doi: 10.1038/sj.onc.1201252. [DOI] [PubMed] [Google Scholar]
- 8.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Chen PL, Subramanian T, Chinnadurai G, Tomlinson G, Osborne CK, Sharp ZD, Lee WH. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J Biol Chem. 1999;274:11334–11338. doi: 10.1074/jbc.274.16.11334. [DOI] [PubMed] [Google Scholar]
- 10.Chinnadurai G. CtIP, a candidate tumor susceptibility gene is a team player with luminaries. Biochim Biophys Acta. 2006;1765:67–73. doi: 10.1016/j.bbcan.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci USA. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 13.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 14.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 15.Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer. 2006;6:382–391. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 16.Lafarge S, Montane MH. Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucl Acids Res. 2003;31:1148–1155. doi: 10.1093/nar/gkg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reidt W, Wurz R, Wanieck K, Chu HH, Puchta H. A homologue of the breast cancer-associated gene BARD1 is involved in DNA repair in plants. EMBO J. 2006;25:4326–4337. doi: 10.1038/sj.emboj.7601313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 19.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 20.Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han P, Li Q, Zhu YX. Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell. 2008;20:1482–1493. doi: 10.1105/tpc.108.058867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon CS, Chen C, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 24.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 26.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 31.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 34.Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Makaroff CA, Ma H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell. 2003;15:1281–1295. [PMC free article] [PubMed] [Google Scholar]
- 36.Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 37.Emerling BM, Bonifas J, Kratz CP, Donovan S, Taylor BR, Green ED, Le Beau MM, Shannon KM. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21:4849–4854. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 38.Klinge B, Uberlacker B, Korfhage C, Werr W. ZmHox: a novel class of maize homeobox genes. Plant Mol Biol. 1996;30:439–453. doi: 10.1007/BF00049323. [DOI] [PubMed] [Google Scholar]