Abstract
At the onset of mitosis, microtubules form a bipolar spindle around the prophase nucleus. TPX2 is phosphorylated during mitosis and acts as a spindle assembly factor that nucleates microtubules in the close vicinity of chromosomes, independent of the centrosomes. Furthermore, it activates the kinase Aurora A and targets the Xenopus kinesin-like protein 2 to spindle poles. We have characterized the plant orthologue of TPX2 that possesses all identified functional domains of its animal counterpart. Moreover, we have demonstrated that it is exported before nuclear envelope breakdown and that its activity around the nuclear envelope is essential for prospindle assembly. Here, we compare the sequences of several characterized TPX2 domains, allowing us to define TPX2. We propose that true TPX2 orthologues share simultaneously all these conserved domains and that other proteins possessing only some of these functional blocks may be considered as TPX2-related proteins.
Key words: mitosis, microtubules, spindle assembly, TPX2 signature, targeting domains, Prosite motifs, evolution
Introduction
The Targeting Protein for Xklp2 (TPX2) has been well characterized in the animal kingdom. TPX2 was first shown to target the plus-end directed kinesin Xklp2 to the minus-ends of spindle microtubules. Its main function, however, is to nucleate microtubules around chromosomes in a RanGTP-dependent manner.1 This early mitotic event, essential for spindle assembly is controlled by importins.2 Two amino acids located in an NLS region of TPX2, conserved between Xenopus and human, play a crucial role in importin α binding and nuclear targeting of the protein. Their mutation, by abolishing importin α interaction, causes TPX2 to become constitutively active in Xenopus M-phase egg extract.3 Furthermore, TPX2 is a microtubule associated protein (MAP) and may participate in microtubule dynamics, bundling and/or spindle pole focusing through its ability to bind microtubules.3 And last but not least, the N-terminal domain of TPX2 binds and activates Aurora A kinase.4
One putative orthologue of TPX2 found in the Arabidopsis thaliana genome shares all these functional domains with its animal counterpart (Table 1).5 Moreover, the protein is exported from the nucleus through the activity of a nuclear export signal (NES) during late G2 phase. Inhibition of the TPX2 function in the cytoplasm blocks prospindle formation and cell cycle progression. These data suggest that in higher plants, TPX2 nucleates perinuclear microtubules leading to the formation of a prospindle, and then may act as a spindle microtubule nucleator and stabilizer.
Table 1.
TPX2, its motifs and proteins sharing homology with some motifs
| Swiss prot accession No | Protein length | organism | name | Aurora A binding motif + central MBD | TPX2 signature motif | C-terminal MBD and Xklp2 binding motif |
| Q6NUF4 | 716 AA | X. laevis | XlTPX2 | + | + | + |
| Q9ULW0 | 747 AA | H. sapiens | HsTPX2 | + | + | + |
| A2APB8 | 745 AA | M. musculus | MmTPX2 | + | + | + |
| Q5ZIC6 | 746 AA | G. gallus | GgTPX2 | + | + | + |
| B3RVY1 | 748 AA | T. adhaerens | TaTPX2 | + | + | + |
| A9V4Q7 | 740 AA | M. brevicolis | MbTPX2 | + | + | + |
| A8J7A6 | 609 AA | C. reinhardtii | CrTPX2 | + | + | + |
| A3BK56 | 767 AA | O. sativa | OsTPX2 | + | ++ | + |
| A5B0H4 | 941 AA | V. vinifera | VvTPX2 | + | ++ | + |
| Q56XS5 | 758 AA | A. thaliana | AtTPX2 | + | ++ | + |
| Q95XR5 | 507 AA | C. elegans | TPXL-1 | Different | − | − |
| A7QGV1 | 174 AA | V. vinifera | TPX2-related | + | − | − |
| Q0WVY6 | 306 AA | A. thaliana | TPX2-related | + | − | − |
| At5g15510 | 497 AA | A. thaliana | TPX2-related | − | − | + |
| Q9C7U3 | 488 AA | A. thaliana | TPX2-related | − | − | + |
| Q5XVC4 | 488 AA | A. thaliana | TPX2-related | − | − | + |
| Q84ZT9 | 202 AA | A. thaliana | WVD2 | − | − | + |
| A5X7Z1 | 176 AA | Populus tremula × P. tremuloides | PttMAP20 | − | − | + |
| At5g44270 | - | A. thaliana | TPX2 pseudogene | 0 | 0 | 0 |
Note: having a particular domain does not mean the actual binding has been proven. Only proteins with all four motifs should be considered true TPX2s.
Various proteins sharing some similarities with TPX2 have been described previously.6–8 These proteins from Caenorhabditis elegans, Arabidospis thaliana and Populus tremula × P. tremuloides share either an N-terminal Aurora A binding site or a C-terminal consensus sequence characterized as a putative binding domain to Xklp2 (PFAM06886) (Table 1). This suggests that TPX2 may belong to a larger protein family or may be built up of a number of joined functional domains. In the latter case, it can be assumed that true TPX2 orthologues must share all these building blocks and that related proteins may possess only some of them. To discriminate between true TPX2 and TPX2 related proteins present in eukaryotes, we determined several amino acid sequence signatures that represent the different functional identities. We show that only one TPX2 gene is expressed per genome and that numerous related proteins contain either the conserved C-terminal microtubule binding domain—and belong therefore to a MAP family—or a conserved Aurora binding domain.
TPX2 Functional Signature
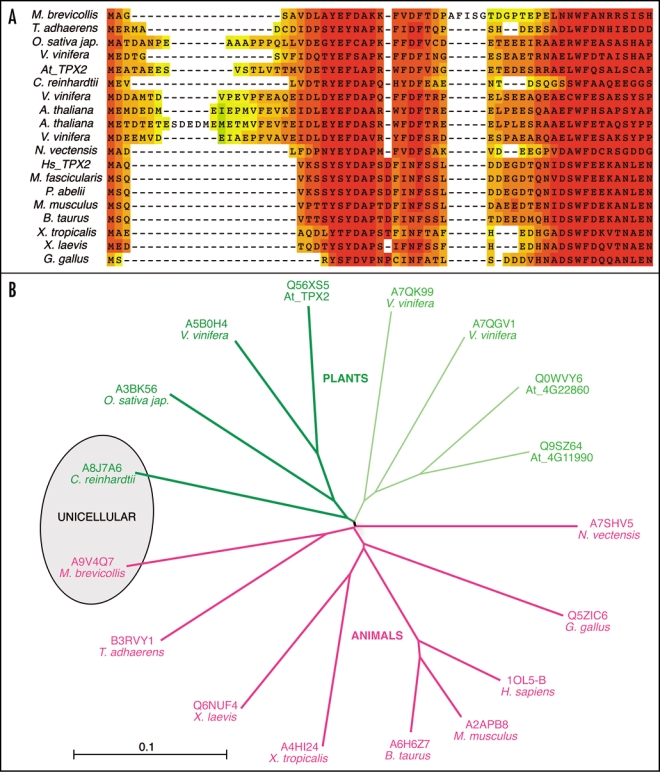
The best characterized domain of TPX2 is located at its N-terminus and mediates its interaction with Aurora A, leading to kinase activation. This interaction plays a central role in spindle assembly.9,10 Interestingly, this mechanism of activation of Aurora A is highly specific as TPX2 does not activate the closely related kinase Aurora B in which a single amino acid substitution in the catalytic domain is sufficient to prevent its binding to TPX2.11 The crystallographic structure of the complex formed between the catalytic domain of Aurora A and the N-terminal part of human TPX2 allowed to define precisely the critical residues involved in direct interactions.4 Based on a multiple sequence alignment analysis among the N-terminal sequences of animal TPX2 proteins and Arabidopsis TPX2, we determined a consensual signature for this TPX2 domain as Y-[EST]-[YF]-X-[VA]-X(4,5)-[ND]-F-X(11,19)-W-F. Using this Aurora binding signature we performed a Prosite12 search on all available databases present on http://expasy.org and identified 41 eukaryote sequences. A partial T-coffee13,14 representation of 19 of the sequences found in a wide range of organisms is shown in Figure 1A. 17 representative sequences were then used to build a relationship tree (Fig. 1B). The putative Aurora binding proteins are expressed in unicellular and non chordate multicellular organisms, early chordates, vertebrates, as well as in mosses and higher plants.
Figure 1.
Identification of proteins sharing the TPX2 Aurora binding motif. (A) T-coffee sequence comparison of 19 N-terminal domains of representative proteins capable of interacting with Aurora A type kinases. The region shown corresponds to the one available in the 3D PDB entry 1OL5-B (human TPX2). The two red blocks are the essential domains that fit within the double Aurora A grooves. (B) Derived distance tree between 17 proteins sharing a putative Aurora A type binding domain. Bold branches highlight true TPX2 proteins and thin ones correspond to plant TPX2-related proteins. The sequences of Macaca fascicularis (Q4R8N4) and Pongo abelii (Q5RAF2) are undistinguishable from the human TPX2 sequence and are therefore not added to the tree.
As plants have three Aurora related kinases that currently cannot be assigned to any of the three Aurora families, we used a molecular modeling approach based on the crystal structure of the human Aurora A—human TPX2 complex to predict which of the three plant Aurora kinases could interact with the N-terminal segment of AtTPX2. We found that the AtTPX2 N-terminal segment could interact with either Arabidopsis AURORA 1 or 2 but not with Arabidopsis AURORA 3.5 This suggests that indeed AtTPX2 interacts with and may activate an Aurora kinase in plants and that like in vertebrates the different members of this family in plants have different mechanism of activation.
The other functional domain of TPX2 lies in its C-terminal half that was shown to be sufficient to promote microtubule nucleation in Xenopus egg extract.15 Among the 41 sequences of TPX2 orthologues that we examined, 37 shared a sequence in this region that we defined as the specific TPX2 signature: R-X-R-[PAS]-X(3)-K-X(5)-E-X-E-[EMQ]-X(3,8)-[HPYLF]-[KPQ]-F-[KPQ]-A (Fig. 1B, bold lines). A blast search with this TPX2 signature revealed moreover that it is found twice in all plant sequences.5 Interestingly, the screen also identified four plant proteins having a putative Aurora A binding domain but no TPX2 signature. This group of proteins dichotomized in our T-coffee based tree, suggesting that they may correspond to different Aurora activators or substrates (Fig. 1B). Strikingly, no proteins from fungus, insect or worm sequence appeared in our searches, suggesting that during evolution these organisms have diverged with regard to TPX2. Altogether, this strengthens the idea that the motif we characterized as a TPX2 signature is particularly relevant.
Microtubule Binding Activity
Two domains of human, Xenopus and Arabidopsis TPX2 may interact with microtubules.15,5 One of them involves the C-terminal domain of TPX2, the other a more central domain of the protein. For AtTPX2, we do not know whether this binding is direct or indirect. Nevertheless, only the central domain of Xenopus15 and human TPX216 was able to nucleate microtubules and to bind purified taxol stabilized microtubules in vitro. As the C-terminal domain of XlTPX2 has been only shown to colocalize with spindle microtubules in a Xenopus mitotic extract, it suggests that this interaction requires additional factors.15 To characterize these microtubule binding activities, we first used the C-terminal sequence L-X(4)-R-A-X-E-R-X-[ED]-[FL]-[ED]-X(4)-E from AtTPX2 as a template to identify other Arabidopsis proteins sharing such structural similarities. 5 sequences were found in various loci on chromosomes 3 and 5 (Table 1). One of these (at5g44270) corresponds to a TPX2 pseudogene. Indeed, although this chromosome region bears a very strong homology with all the TPX2 exons and is transcribed, the generated messenger is mis-spliced and therefore cannot be translated normally. All 5 proteins are shorter than AtTPX2 and do not possess the Aurora binding domain. They may just have a conserved TPX2-specific microtubule binding domain (MBD) and therefore cannot be considered as true TPX2 proteins. When expressed as GFP fusions in bombarded BY-2 cells, we showed that these proteins labeled interphase microtubules, confirming their ability to bind microtubules in vivo.
Using the same T-coffee comparison approach, we defined another sequence for the C-terminal domain of TPX2: [VFL]-X-L-X(4)-R[AV]-X-[EK]-R-X-X-[FL]-[EDNK]-X(32)-R-X(6)-A to find other MAPs. Even though the stringency of this motif may be too high to detect all related sequences, it did not provide false positives in our screen. When this motif was used in a Prosite search, once again, all previously characterized TPX2 sequences came out. Moreover, as found earlier with the Aurora binding signature, additional sequences that did not share further similarities with TPX2 appeared, suggesting that the microtubule binding activity has been spread among many proteins as a functional unit (i.e., MAP20 in the poplar genome, see Table 1). Homologues of the poplar PttMAP20 have been identified in other plant genomes.17 However, the role of PttMAP20 in secondary cell wall synthesis highly suggests that it is far from playing TPX2 functions. It may more likely stabilize cortical microtubules along which cellulose synthase rosettes are guided as they synthesize parallel microfibrils in the cell wall.18 Members of the WVD2/WDL family also have an MBD related to the TPX2 C-terminal domain that seems to be involved in cortical microtubule binding.7
The second MBD of AtTPX2 involves a centrally located sequence downstream from the Aurora binding site. It also contains an NLS that targets the protein to the nucleus. Its MBD activity was therefore only observed after mutation of the NLS basic amino acids. Like the AtTPX2 C-terminal fragment, the four full-length Arabidopsis and Vitis TPX2-related proteins shown in Figure 1B (regular lines), bind microtubules when transiently expressed as fusion proteins with GFP upon bombardment of their corresponding plasmid. None of our in silico analysis of MBD signatures returned the C. elegans TPXL-1 protein, suggesting strongly that this protein is not a true TPX2 orthologue.19
Conclusions
TPX2 proteins have first been functionally defined as necessary to target the plus-end directed motor Xklp2 to the mitotic spindle poles although no direct interaction between these proteins has been demonstrated.1 Concerning such a putative Xklp2 binding activity, the consensual PF06886 sequence of ≈60AA described in the literature highly overlaps the C-terminal MBD. When it is used to screen bioinformatic libraries,17 it does not specifically sort out TPX2 proteins. Currently, we do not know if AtTPX2 binds any specific kinesin motor. In any case, our data strongly argue in favor of one unique TPX2 gene per genome, sharing the functions described for Xenopus, mouse and human TPX2. All the other TPX2-related genes identified so far possess either the Aurora binding domain and/or one microtubule binding domain. This suggests that proteins are built up of functional units that spread through the genome and through evolution. We suggest to replace the FPAM 06886 sequence, currently used to characterize TPX2, by that one we demonstrated to be a specific TPX2 signature.
Acknowledgements
This work was in part supported by the Centre National de la Recherche Scientifique (CNRS), partenariat Hubert Curien (PHC)-Van Gogh and PHC-Picasso collaboration grants by Egide to A.C. Schmit. The Inter-Institute confocal microscopy platform was co-financed by the CNRS, the Université Louis Pasteur, the Région Alsace, the Association de la Recherche sur le Cancer and the Ligue Nationale contre le Cancer. This work was also partially supported by a VENI grant and a Van Gogh collaboration grant, both by the Netherlands Organization for Scientific Research (NWO), to J.W. Vos. The work of L. Pieuchot was granted by the “Ministère de l'Enseignement Supérieur et de la Recherche”. I. Vernos was supported by Spanish grants (HF-2006-0067; BFU2006-04694 and CSD2006-00023) and by the EU grant MRTN/CT2004 512348.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7409
References
- 1.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel Xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 3.Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 5.Vos JW, Pieuchot L, Evrard JL, Janski N, Bergdoll M, de Ronde D, Perez LH, Sardon T, Vernos I, Schmit AC. The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell. 2008 doi: 10.1105/tpc.107.056796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozlu N, Srayko M, Kinoshita K, Habermann B, O'toole ET, Muller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Perrin RM, Wang Y, Yuen CY, Will J, Masson PH. WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 2007;49:961–971. doi: 10.1111/j.1365-313X.2006.03015.x. [DOI] [PubMed] [Google Scholar]
- 8.Rajangam AS, Kumar M, Aspeborg H, Guerriero G, Arvestad L, Pansri P, Brown CJ, Hober S, Blomqvist K, Divne C, Ezcurra I, Mellerowicz E, Sundberg B, Bulone V, Teeri TT. MAP20, a microtubule-associated protein in the secondary cell walls of Populus tremula L. x tremuloides Michx is a target of the cellulose synthesis inhibitor, 2,6-dichlorobenzonitrile. Plant Physiol. 2008 doi: 10.1104/pp.108.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sardon T, Peset I, Petrova B, Vernos I. Dissecting the role of Aurora A during spindle assembly. EMBO J. 2008;27:2567–2579. doi: 10.1038/emboj.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardon T, Peset I, Petrova B, Vernos I. Dissecting the role of Aurora A during spindle assembly. EMBO J. 2008;27:2942. doi: 10.1038/emboj.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayliss R, Sardon T, Ebert J, Lindner D, Vernos I, Conti E. Determinants for Aurora-A activation and Aurora-B discrimination by TPX2. Cell Cycle. 2004;3:404–407. [PubMed] [Google Scholar]
- 12.Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 13.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucl Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirot O, Suhre K, Abergel C, O'Toole E, Notredame C. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucl Acids Res. 2004;32:37–40. doi: 10.1093/nar/gkh382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet S, Sardon T, Zimmerman T, Wittmann T, Pepperkok R, Karsenti E, Vernos I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol Biol Cell. 2004;15:5318–5328. doi: 10.1091/mbc.E04-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trieselmann N, Armstrong S, Rauw J, Wilde A. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J Cell Sci. 2003;116:4791–4798. doi: 10.1242/jcs.00798. [DOI] [PubMed] [Google Scholar]
- 17.Rajangam AS, Hongqian Y, Teeri TT, Arvestad L. Evolution of a domain conserved in microtubule-associated proteins of eukaryotes. Comput Biol Chem. 2008;1:1–18. doi: 10.2147/aabc.s3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 19.Karsenti E. TPX or not TPX? Mol Cell. 2005;19:431–432. doi: 10.1016/j.molcel.2005.08.002. [DOI] [PubMed] [Google Scholar]