Abstract
Reversible posttranslational modification of proteins by the action of small ubiquitin-like modifier (SUMO) peptide (sumoylation) has been known to participate in various biological processes in eukaryotes. However, much less is known about the role of sumoylation in plants. In our recent paper to which we write this Addendum, we show that loss of SIZ1, a SUMO E3 ligase, results in a highly increased SA-mediated defense signaling through a PAD4-dependent pathway. This signaling leads to constitutively expressed pathogen related (PR) genes and to increased disease resistance to a virulent bacterial pathogen. These findings significantly increase our understanding of the role of sumoylation in the plant defense system.
Key Words: plant innate immunity, salicylic acid (SA), small ubiquitin-like modifer (SUMO), plant-pathogen interactions
Sumoylation is a posttranslational modification process that covalently conjugates the small ubiquitin-like modifier (SUMO) protein to the K (lysine) residues located at binding sites of sumoylation target proteins.1,2 The SUMO conjugation/de-conjugation process occurs in a sumoylation cycle that is mediated by E1-activating protein, E2-conjugation protein, E3-ligase and SUMO protease.2 The 3-D structure of SUMO peptide is highly similar to the ubiquitin peptide that affects many biological processes by mediating protein-degradation. Even though sumoylation and ubiquitination may, in some instances, affect the same target protein, the biological function of sumoylation can be substantially different from ubiquitination.3 In yeast and animals, SUMO is linked to numerous subcellular processes such as cell cycle progression, DNA repair, nucleocytoplasmic trafficking, subnuclear targeting, ubiquitination antagonism, and transcriptional regulation.2–5
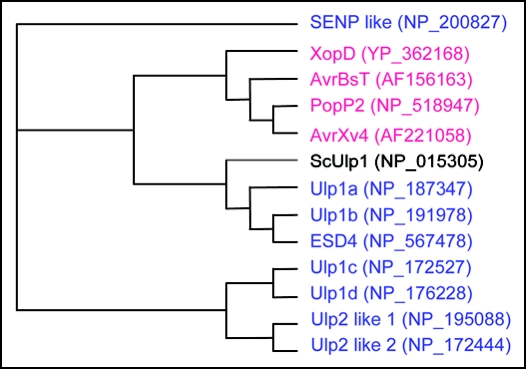
Although there are few examples of SUMO function in plants, in vitro and in planta biochemical evidence has established the existence of an Arabidopsis SUMO sumoylation/de-sumoylation system and this system appears to play an important role in various plant environmental responses including heat shock, oxidative stress, ABA signaling, flowering, and phosphate deficiency.6–10 In addition to these responses, it has long been speculated that SUMO could have an important role in plant defense against pathogens. Even though bacteria do not contain a sumoylation/de-sumoylation system, they have proteins that are structurally similar to SUMO proteases (Fig. 1). The Xanthomonase campestris effectors, XopD and AvrXv4 migrate to the plant nucleus after injection by the type III secretion system, and have SUMO protease activity.11–13 Disruption of SUMO isopeptidase activity of AvrXv4 prevents the hypersensitive response in tabacco.13 There are three specific SUMO genes (SUMO 1, 2, 3) that encode SUMO peptides that are slightly different in size and sequence from each other. Interestingly, in vitro analyses reveal that XopD can cleave all three SUMO conjugates. However, Arabidopsis SUMO proteases ULP1C, ULP1D, and ESD4 cleave SUMO1, 2-but not SUMO3-conjugates.14 These results indicate that the SUMO conjugation/de-conjugation cycle plays some role in the plant innate immunity response system. However, to this point, direct evidence of any details for the role of SUMO in plant defense responses have been lacking.
Figure 1.
Phylogenic analysis of SUMO protease-like proteins from plant pathogens (red), Arabidopsis (blue), and yeast (black). Proteins were grouped into a phylogram using the Clustal W server at the http://www.ebi.ac.uk/clustalw/.
In our study, we examined the role of SIZ1, a SUMO E3 ligase, in plant pathogen defense. The siz1 mutant plants have a reduced growth phenotype, overexpress PR genes and exhibit resistance to the pathogen Pseudomonas syringae pv. tomato (Pst). These characteristics are seen in other mutant plants with high levels of salicylic acid (SA).15 As we expected, endogenous SA levels in siz1 plants are elevated. When the nahG gene, which encodes a salicylic acid hydroxylase, is introduced into siz1 mutant plants, a decreased SA level, and a reversal of the other altered phenotypes to wild-type results.15 SIZ1 controls PR gene expression and disease resistance through an epistatic interaction with PAD4. Further, Pseudomonas resistance of siz1 plants is mediated specifically by a EDS1/PAD4-dependent TIR-NBS-LRR-type R gene pathway.15 Based on these observations, we concluded that SIZ1 SUMO E3 ligase negatively regulates SA-mediated pathogen defense signaling through a PAD4-dependent process in Arabidopsis.
We have recognized PAD4, EDS1, and SAG101 as possible targets of SIZ1. These proteins have conserved SUMO attachment motifs and regulate SA biosynthesis. Also, because the EDS1/PAD4-mediated specific R gene pathway is hyperactivated in siz1 plants, it is possible that specific R protein(s) that regulate expression of PAD4/EDS1 might be direct or indirect targets of SIZ1. Recently, it has been reported that Ralstonia. solanacerum PopP2, a type III effector protein that structurally resembles SUMO protease, physically interacts with RRS1, a TIR-NBS-LRR type R protein, from A. thaliana (Fig. 1).16 The identification of specific R proteins that are modified by sumoylation/de-sumoylation would reveal a new important regulatory mechanism that controls plant-pathogen interactions.
Acknowledgements
This work was supported by grants from the Plant Diversity Research Center of the 21st Century Frontier Research Program of MOST (PF0330401-00), the Environmental Biotechnology National Core Research Center Project of KOSEF (R15-2003-01201002-00).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/3867
References
- 1.Verger A, Perdomo J, Crossley M. Modification with SUMO. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Gill G. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 4.Freiman RN, Tjian R. Regulating the regulators: Lysine modifications make their mark. Cell. 2003;112:11–17. doi: 10.1016/s0092-8674(02)01278-3. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Viestra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 7.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. SIZ1 Small Ubiquitin-Like Modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;12:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lois LM, Lim CD, Chua NH. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 12.Hotson A, Chosed R, Shu H, Orth K, Mudgette MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in plants. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 13.Roden J, Eardley L, Hotson A, Cao Y, Mudgette MB. Characterization of the Xanthomonase AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microb Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 14.Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 16.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marcho Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]