Abstract
We have previously reported that Al-induces citrate and malate efflux from P-sufficient and P-deficient plants of rape (Brassica napus L.) and that P-deficiency alone could not induce this response. Further investigation showed that the transcript of two genes designated BnALMT1 and BnALMT2 is accumulated in roots by Al-treatment. Transgenic tobacco cells (Nicotiana tabacum) and Xenopus laevis oocytes expressing the BnALMT1 and BnALMT2 proteins released more malate than control cells in the presence of Al, indicating that the BnALMT genes encode an Al-activated malate transporter. The transgenic tobacco cells exposed to toxic level of Al grew better than control cells indicating that the genes can enhance Al-resistance of plant cells. In this study we showed the subcellular localization of BnALMT1 fused to the green fluoresce protein (GFP). The BnALMT1:: GFP construct was transiently expressed in protoplasts prepared from Arabidopsis leaves using the polyethylene glycol (PEG) method. The result showed that the BnALMT1 protein is localized in the plasma membrane. This provides further evidence that the BnALMT proteins facilitate the transport of malate across the plasma membrane (PM).
Key Words: Brassica napus, Aluminum toxicity, malate efflux, BnALMT, GFP, PM, Nicotiana tobacum, Xenopus laevis oocytes, Al-tolerance
Aluminium toxicity limits crop production on the acidic soils that comprise over 40% of the world's potentially arable lands.1 On such soils, P-deficiency is commonly reported together with Al-toxicity.2 When the soil pH falls bellow 5, Al is solublized into toxic forms and then inhibits root growth and functions.
The release of organic anions such as malate, citrate and oxalate from plant roots is believed to be a mechanism of Al resistance.3–8 Induction of organic anions efflux was also reported in P-deficient rape,9 white lupin10 and alfalfa.11 However, in our previous investigation we did not observe any induction of organic anion efflux under P-deficient condition,12 but we have shown that Al-induced efflux is more pronounced in P-sufficient plants of rape.
The Al resistance in wheat relies on the Al-dependent efflux of malate anions from the root apices and a strong correlation exists between relative Al resistance of different genotypes and the capacity for malate efflux.4,13 Pharmacological and electrophysiological studies suggest that malate efflux is facilitated by anion channels and this is consistent with the electrochemical gradient for anions across the plasma membrane.14,15
The TaALMT1 gene which controls the Al-dependent efflux of malate was first isolation from wheat.16 TaALMT1 expression in Al-resistant genotypes of wheat is 5 to 10-fold higher than in Al-sensitive genotypes.16,17 Heterologous expression of TaALMT1 in cultured-tobacco cells, Xenopus oocytes, and intact rice and barley plants conferred an Al-activated malate efflux and increased the Al-tolerance of tobacco suspension cells and intact barley plants.16,18 Recently AtALMT1, one of the homologues of TaALMT1 in Arabidopsis, has been implicated in an Al-resistance mechanism which also relies on malate release.19 In an effort to elucidate the molecular mechanism underlying the Al-induced malate efflux in rape we have cloned two homologues of the TaALMT1 gene, designated BnALMT1 and BnALMT2. The transcript of BnALMT1 and BnALMT2 was accumulated in the root, not shoot by Al in a concentration dependant manner.20 This is similar to AtALMT119 but contrasts with TaALMT1 which is constitutively expressed in wheat and unaffected by Al treatment.16 Our result also showed that neither BnALMT1 nor BnALMT2 is induced by P-deficiency.20 Therefore, we conclude that P-deficiency does not induce both BnALMT transcript accumulation and malate efflux in rape.12,20
Overexpression of the BnALMT proteins in tobacco cells and Xenopus oocytes conferred an Al-induced malate efflux. Citrate was not released from the transgenic cells, indicating that the BnALMT proteins are specific for malate transport. Hence, we suggest that other proteins might be involved in facilitating the Al-induced citrate efflux from wild type plants of rape.12 In line with this, other researchers have identified an Al tolerance locus in sorghum that segregates with citrate release from roots.21 If such genes are identified, future studies can examine whether similar genes are involved in citrate release from rape. Furthermore, hetrologous expression of the BnALMT proteins enhanced the Al-resistance of the transgenic tobacco cells. It is worthwhile therefore to overexpress the BnALMT proteins in other economically important crops in order to maximize crop productivity on the acidic soils where Al-toxicity is a major problem.
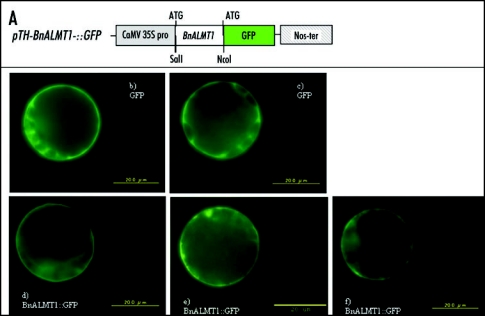
The present study focused on the subcelluar localization of BnALMT1 protein. For this purpose, BnALMT1 was cloned into pTH-2 vector at the SalI and NcoI site in frame with the green florescencet protein (GFP) fused to the C-terminal site (Fig. 1a). The construct under the control of 35S promoter and nopaline synthase (nos) terminator was transiently expressed in Arabidopsis leaf protoplast using the polyethylene glycol (PEG) method as previously described.22 Transformed protoplasts were examined using a fluorescent microscope (BX61 equipped with a sectioning device DSU, Olympus) with a B-excitation filter set (for GFP).
Figure 1.
Expreesion of BnALMT1::GFP fusion protein in Arabidosis (ecotype Colombia) leaf protoplast. BnALMT1 was cloned into pTH-2 at the SalI and NcoI site in frame with the GFP on the C-terminal site to yield a construct pTH-BnALMT1::GFP (A). The construct was transiently expressed in Arabidopsis leaf protoplast using the PEG method of transformation (D–F). For comparison, pTH-GFP was used as control (B and C). Representative images are presented.
The result showed that the fluorescent signal of the BnALMT1::-GFP construct was more confined to the plasma membrane (Figs. 1d, 1e and 1f) unlike the control (expressing only the GFP) which seems not to be restricted to the PM but also observed in the cytoplasm (Figs. 1b and 1c). Similarly, the TaALMT1 protein is localized in the plasma membrane.23 Therefore, we conclude that the BnALMT proteins are localized in the plasma membrane and facilitate the transport of malate out of plant cells exposed to toxic Al.
The responses of rape genotypes to Al-stress have not been well understood. Hence, future study will examine these responses and whether genotypic variation in Al-tolerance exists among rape genotypes. Furthermore, the polymorphism in the two BnALMT1 genes can be studied to generate a molecular marker which can be used to map the location of the two genes on the rape genome. If significant variations are observed among the parental lines, the marker can be used to study if Al-tolerance segregates with either of the BnALMT genes.
Acknowledgements
We thank Ms. Chieko Hattori for technical support. This research was supported by a postdoctoral fellowship awarded by the Japan Society for the Promotion of Science (to A.L.).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/3868
References
- 1.von Uexkuell HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- 2.Clark RB. Effect of aluminium on growth and mineral elements of aluminium tolerant and aluminium intolerant corn. Plant Soil. 1977;47:653–662. [Google Scholar]
- 3.Miyasaka SC, Buta JG, Howell RK, Foy CD. Mechanism of aluminium tolerance in snap- beans. Root exudates of citric acid. Plant Physiol. 1991;96:737–743. doi: 10.1104/pp.96.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delhaize E, Ryan PR, Randall P. Aluminium tolerance in wheat (triticum aestivum L.). II. Aluminium-stimulated excretion of malic acid from root apices. Plant Physiol. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellet DM, Grunes DL, Kochian LV. Organic acid exudation as an aluminium-tolerance mechanism in maiz (Zea mays L.) Planta. 1995;196:788–795. [Google Scholar]
- 6.Ryan PR, Delhaize E, Randall PJ. Characterization of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta. 1995;196:103–110. [Google Scholar]
- 7.Ma JF, Zheng SJ, Matsumoto H, Hiradate S. Detoxifying aluminum with buckwheat. Nature. 1997;390:569–570. [Google Scholar]
- 8.Zheng SJ, Ma JF, Matsumoto H. High aluminium resistance in buckwheat I, Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998;117:745–751. doi: 10.1104/pp.117.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffland E, Findenegg GR, Nelemans JA. Utilization of rock phosphate by rape. Plant Soil. 1989;113:155–160. [Google Scholar]
- 10.Gardner WK, Barber DA, Parbery DG. The acquisition of phosphorus by Lupinus albus L. III. The probable mechanism by which phosphorus movement in the soil root interface is enhanced. Plant Soil. 1983;70:107–124. [Google Scholar]
- 11.Lipton D, Blanchar R, Blevins D. Citrate, malate and succinate concentration in exudates from P sufficient and P stressed Medicago sativa L. seedlings. Plant Physiol. 1987;85:315–317. doi: 10.1104/pp.85.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ligaba A, Shen H, Shibata K, Yamamoto Y, Tanakamaru S, Matsumoto H. The role of phosphorus in aluminum-induced citrate and malate exudation in rape (Brassica napus L.) Physiol Plant. 2004;120:575–584. doi: 10.1111/j.0031-9317.2004.0290.x. [DOI] [PubMed] [Google Scholar]
- 13.Ryan PR, Delhaize E, Randall PJ. Malate efflux from root apices and tolerance to aluminium are highly correlated in wheat. Aust J Plant Physiol. 1995;22:531–536. [Google Scholar]
- 14.Ryan PR, Skerrett M, Findlay GP, Delhaize E, Tyerman S. Aluminum activates an anion channel in the apical cells of wheat roots. Proc Natl Acad Sci USA. 1997;94:6547–6552. doi: 10.1073/pnas.94.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W-H, Ryan PR, Tyerman S. Malate-permeable channels and cation channels activated by aluminum in the apical cells of wheat roots. Plant Physiol. 2001;125:1459–1472. doi: 10.1104/pp.125.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 17.Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, et al. Molecular characterization and mapping of ALMT1, the aluminium- tolerance gene of bread wheat (Triticum aestivum L.) Genome. 2005;48:781–791. doi: 10.1139/g05-054. [DOI] [PubMed] [Google Scholar]
- 18.Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoekenga OA, Maron LG, Cançado GMA, Piñeros MA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Koyama H, Kochian LV. AtALMT1 (At1g08430) is a novel, essential factor for aluminum tolerance in Arabidopsis thaliana and encodes an aluminum-activated malate transporter. Proc Natl Acad Sci USA. 2006;103:9734–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligaba A, Katsuhara M, Ryan RP, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from Brassica napus L. encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV. Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the poaceae. Genetics. 2004;167:1905–1914. doi: 10.1534/genetics.103.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H. Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1) Plant Cell Physiol. 2005;46:812–816. doi: 10.1093/pcp/pci083. [DOI] [PubMed] [Google Scholar]