Abstract
Nitrate transporters are important for nitrogen acquisition by plants and in algae some require two gene products, NRT2 and NAR2, for function. The NRT2 family was already described and the recent identification of a family of the NAR2-type genes in higher plants showed that there was a homologue in Arabidopsis, AtNAR2.1. Using heterologous expression in yeast and oocytes we showed that the two Arabidopsis AtNRT2.1 and AtNAR2.1 proteins interacted to give a functional high affinity nitrate transport system (HATS). The gene knock out mutant atnar2.1-1 is deficient specifically for HATS activity and the resulting growth phenotype on low nitrate concentration is more severe than for the atnrt2.1-1 knock out mutant. Physiological characterisation of the plant N status and gene expression revealed a pattern that was characteristic of severe nitrogen deficiency. Consistent with the down regulation of AtNRT2.1 expression, the atnar2.1-1 plants also displayed the same phenotype as atnrt2.1 mutants in lateral root (LR) response to low nitrate supply. Using atnar2.1-1 plants constitutively expressing the NpNRT2.1 gene, we now show a specific role for AtNAR2.1 in LR response to low nitrate supply. AtNAR2.1 is also involved in the repression of LR initiation in response to high ratios of sucrose to nitrogen in the medium. Therefore the two component system itself is likely to be involved in the signaling pathway integrating nutritional cues for LR architecture regulation. Using a green fluorescent protein-NRT2.1 protein fusion we show the essential role of AtNAR2.1 for the presence of AtNRT2.1 to the plasma membrane.
Key Words: high affinity nitrate transport, nitrate transporter, nitrate signalling, root growth
Introduction
Nitrate uptake by plants is a fundamental aspect of their nutrition and several families of transporters have been identified.1,2 For one of these families, the NRT2s, a second protein has been shown to be necessary for the nitrate transport function in Xenopus oocytes3,4 and in planta using gene knock-out mutants.5,6 Earlier research using the green alga Chlamydomonas had identified that a second gene was important for high affinity nitrate transport and it was named Nar2.7 Now it has been shown that the interaction of the two components actually occurs between the two proteins.5 Gene knock-out Arabidopsis plants that lack normal levels of mRNA for the second protein component, AtNAR2.1, are unable to take up nitrate in the high affinity range at low external concentrations.5,6
When plants are grown on low concentrations of nitrate (0.2 mM), even though the actual amount of nitrate present may be large by volume, the atnar2.1-1 mutant plants cannot access this pool and show all the physiological symptoms of nitrogen (N) deficiency. Wild type plants under these conditions grow normally as they would when supplied with much higher nitrate concentrations. The growth phenotype of atnar2.1-1 plants was so strong when they were grown on low nitrate concentrations (0.2 mM nitrate), that the mutant plants (atnar2.1-1) provide a powerful tool to unravel high affinity nitrate uptake function by other transport proteins. These knock-out mutants also display a root architecture phenotype in responses to low nitrate supply. By crossing the atnar2.1-1 mutant with other plants we may also have a complementation test for the ability of other proteins to substitute for the responses of roots to low nitrate supply.
Lateral root assays to identify if the tobacco NpNRT2.1 gene can compensate for AtNRT2.1 and is AtNAR2.1 essential?
Changes in lateral root (LR) growth are a sensitive and easily measurable response of plants to N limitation.8 Recently these assays were used to demonstrate a central role for the nitrate transporter AtNRT2.1 in the morphological and physiological responses of roots to changes in N supply.9,10 Either disruption of the production of mRNA9 or a point mutation that decreases the transport activity of the AtNRT2.110 was shown to interfere with the response of LR growth to changes in N supply. For both these types of mutants, measurements of 15N-nitrate influx showed that the plants had impaired high affinity uptake, but in addition AtNRT2.1 has some role in LR initiation that was independent of this transport function. The authors suggested a direct stimulatory role for AtNRT2.1 in LR initiation under N-limited conditions9 and a repressive role under high sucrose/ low nitrate conditions.10 When gene knock-out plants that lack either the NRT2.1 (atnrt2.1-1) or NAR2.1 (atnar2.1-1) genes were compared in LR growth assays under low nitrate supply we found that there were some small but significant differences between the two types of mutant.5 For both mutants the response of the root system can be attributed to lower nitrate uptake rates creating a stronger N deprivation status. After 4 d under low nitrate conditions, the atnar2.1-1 mutant cannot sustain an increased growth rate5 while no limitation has been reported for the atnrt2.1-1 mutant.9 This result is probably due to a shortage of N occurring earlier in atnar2.1-1 than atnrt2.1-1 due to a greater limitation of the HATS. In addition, the atnar2.1-1 mutant displays the same inhibition of LR initiation on the newly developed primary root as the atnrt2.1-1 mutant. This phenotype could result from the decreased expression of AtNRT2.1 in the atnar2.1-1 mutant. The root specific constitutive expression of NpNRT2.1 can restore the HATS in the atnrt2.1-1 mutant,11 but not in the atnar2.1-1 mutant5 leading to the conclusion that AtNAR2.1 is essential for a functional HATS. With the RolDNpNRT2.1X atnar2.1-1 plants we generated a tool decoupling the HATS activity and the NRT2.1 gene expression in the roots.5 Therefore we tested the role of the NRT2.1 protein itself in the LR initiation when the plants are transferred to low nitrate supply.
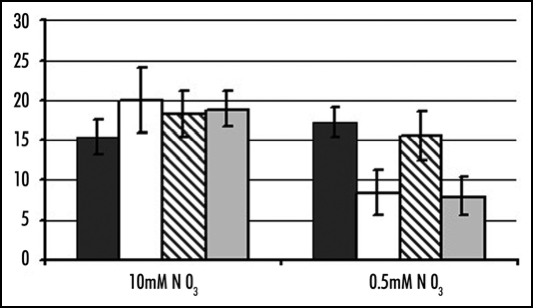
The important result in these assays was the number of new LRs produced on the newly developed primary root.5 Data for RolDNpNRT2.1X atnar2.1-1 plants in comparison to the RolDNpNRT2.1 over-expresser, atnar2.1-1 mutant and wild type are shown (see Fig. 1). On 10 mM nitrate, the LR numbers were not significantly different between the genotypes (Fig. 1), but on 0.5 mM, both RolDNpNRT2.1X atnar2.1-1 and atnar2.1-1 plants had significantly lower numbers of new LRs when compared with wild type and RolDNpNRT2.1 plants. Plants constitutively expressing NpNRT2.1 could not compensate for the lack of AtNAR2.1 for the LR initiation response during N limitation. Therefore, either the tobacco gene cannot functionally replace the Arabidopsis gene AtNRT2.1 for the LR response while it can for the HATS activity,11 or both genes NRT2.1 and AtNAR2.1 are required for the LR response as well as for a functional HATS. The second idea is consistent with the hypothesis that the AtNAR2.1 proteins may be involved in targeting the NRT2.1s proteins to the plasma membrane.5 In this case even if the NpNRT2.1 protein is constitutively expressed in the root, the proteins cannot be properly targeted to the plasma membrane in the atnar2.1-1 background and the LR response cannot be restored.
Figure 1.
Number of new lateral roots on new primary roots. WT (WS, black bars), mutant (atnar2.1-1, white bars), overexpresser (WSRolDNpNRT2.1, cross-hatched bars) and cross (RolDNpNRT2.1X atnar2.1-1, grey bars) plants were grown on vertical agar plates on 10 mM NO3− for 7 d and then transferred to either 0.5 or 10 mM NO3− for a further 6 d (13 d of total growth). Materials and Methods described previously.5 Mean values ± SD of 12 seedling are shown. On 0.5 mM nitrate both atnar2.1-1 and RolDNpNRT2.1X atnar2.1-1 show significantly less new lateral roots when compared with wild type and WSRolDNpNRT2.1. Statistical comparisons of means between treatments or genotypes were performed by the pooled Student's t test using the Sigmaplot software (Systat Software UK, London, UK).
The role of AtNAR2.1 in the repression of LR initiation in response to nutritional cues.
Both proteins AtNRT2.1 and AtNAR2.1 are essential for a functional HATS and also for stimulation of LR development in response to low nitrate. In the next experiment we tested if like AtNRT2.1, AtNAR2.1 was also involved in the repression of LR initiation by nutritional cues.10 Plants were grown on high ratios of sucrose to nitrogen (7.5% sucrose), 1 mM NH4Cl and decreasing concentrations of KNO3 from 5 mM to 0 mM (Fig. 2).
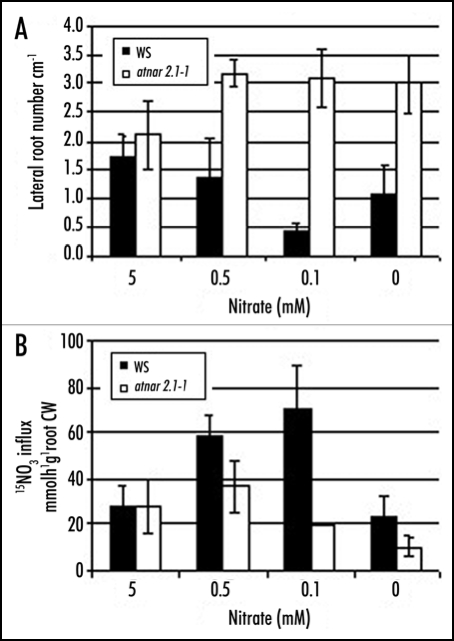
Figure 2.
High sucrose/low nitrate represses lateral root initiation in WT but not atnar2.1. (A) WT (WS, black) and mutant (atnar2.1-1, white) seedlings were grown for 13 days on media containing 7.5% sucrose, 0.1 mM NH4Cl, and various nitrate concentration (0, 0.1, 0.5, 5 mM KNO3).10 KCl was used to balance total salt concentrations between media. The values are means of the number of visible lateral roots per cm of primary root length ± SD of 12 seedlings. (B) Root 15NO3− influx measured at 0.2 mM. The values are means ± SD of three replicates (pooling four to eight plants).
In the high affinity range of nitrate concentrations, LR initiation is gradually repressed in the WT as nitrate concentration is decreasing (5 mM to 0.5 mM and 0.1 mM), while in the atnar2.1-1 mutant, LR initiation is induced and maintained at the same level (Fig. 2A). Nitrate uptake activity is induced in the WT as the concentration is decreasing, but repressed in the atnar2.1-1 mutant (Fig. 2B). As was observed for the atnrt2-1 mutants,10 the LR phenotype is independent of the amount of nitrate absorbed because atnar2.1-1 displays the phenotype even on 0 mM nitrate supply (Fig. 2). Therefore AtNAR2.1 is also essential for the repression of LR initiation in response to nutritional cues.
AtNRT2.1 localisation in the atnar2.1-1 background: a role for AtNAR2.1
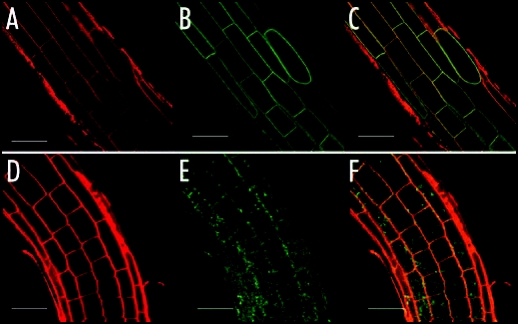
The AtNRT2.1 localisation at the plasma membrane has recently been elucidated using a constitutive expression of the AtNRT2.1-green fluorescent protein (GFP) fusion in the WT background.12,13 In order to investigate the role of AtNAR2.1 in the targeting of AtNRT2.1 to the plasma membrane the 35SAtNRT2.1-GFP transformed plants were crossed with the atnar2.1-1 mutant. The resulting plants are constitutively expressing the AtNRT2.1-GFP protein in the atnar2.1-1 background. While the GFP fluorescence associated with the AtNRT2.1 protein is nicely localised at the plasma membrane in the WT background (Fig. 3B), the fluorescence is lower and diffused throughout the cell in the atnar2.1-1 background (Fig. 3E). In addition, as revealed by l-scanning, the signal is due to an autofluorescence mechanism rather that a true GFP fluorescence. AtNRT2.1 is not properly targeted to the plasma membrane in the atnar2.1-1 mutant. Therefore AtNAR2.1 is likely to be involved in the stability of AtNRT2.1 possibly through the targeting process.
Figure 3.
GFP-NRT2.1 localization in plants expressing the P35S-GFP-NRT2.1 transgene in wild-type background (p43NRT2.1 plants) (A–C) and in the nar2.1-1 mutant background (nar2.1-1/p43NRT2.1 plants) (D–F). Plants were grown in horizontal Petri dishes for 7 days with 1% sucrose. Root cells were stained for 10 min with the endocytic tracer FM4-64, Materials and Methods described previously.12 GFP emission was measured on p43NRT2.1 plants (Ws wild-type transformed with the construct P35S-GFP-NRT2.1) (B), and in nar2.1-1/p43NRT2.1 plants resulting from a cross between the nar2.1-1 mutant and p43NRT2.1 (E). (A) and (D) represent background images of the corresponding plants and (C) and (F) overlays of the GFP and marker images. Bar = 40 µm.
Both AtNRT2.1 and AtNAR2.1 proteins are involved in the regulation of the LR architecture independently of their uptake function. AtNRT2.1 has been suggested to have a role as a nitrate sensor.10 It is likely that AtNAR2.1 also participates in this signaling pathway in a way that is consistent with a role in the targeting of AtNRT2.1 to the plasma membrane.
Final Note
Gene nomenclature-Three names for the same gene.
Naming genes is complicated and one of the two components of the HATS has had three different names. The gene was actually first named WR3 as it was identified as a wound-inducible gene in Arabidopsis.14 When the gene family was found to be important for higher plant nitrate transport4 it was named AtNAR2.1 after the related gene in Chlamydomonas, CrNAR2. Finally, in a recent paper AtNAR2.1 was renamed AtNRT3.1.7
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/3870
References
- 1.Williams LE, Miller AJ. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Ann Rev Plant Physiol Plant Mol Biol. 2001;52:659–688. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- 2.Orsel M, Krapp A, Daniel-Vedele F. Analysis of the NRT2 nitrate transporter family in Arabidopsis: Structure and gene expression. Plant Physiol. 2002;129:886–896. doi: 10.1104/pp.005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou JJ, Fernandez E, Galvan A, Miller AJ. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Letts. 2000;466:225–227. doi: 10.1016/s0014-5793(00)01085-1. [DOI] [PubMed] [Google Scholar]
- 4.Tong Y, Zhou JJ, Li Z, Miller AJ. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005;41:442–450. doi: 10.1111/j.1365-313X.2004.02310.x. [DOI] [PubMed] [Google Scholar]
- 5.Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. Characterisation of a two component high affinity nitrate uptake system in Arabidopsis; physiology and protein-protein interaction. Plant Physiol. 2006;142:1304–1317. doi: 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, Glass ADM. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-Like gene AtNRT3.1. Plant Physiol. 2006;140:1036–1046. doi: 10.1104/pp.105.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quesada A, Galvan A, Fernandez E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994;5:407–419. doi: 10.1111/j.1365-313x.1994.00407.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Forde BG. Regulation of Arabidopsis root development by nitrate availability. J Exp Bot. 2000;51:51–59. [PubMed] [Google Scholar]
- 9.Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Letts. 2001;489:220–224. doi: 10.1016/s0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- 12.Chopin F, Wirth J, Dorbe MF, Lejay L, Krapp A, Gojon A, Daniel-Vedele F. The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane (submitted to Functional Plant Biology) doi: 10.1016/j.plaphy.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A. Control of nitrate uptake activity at the NRT2.1 protein level in Arabidopsis thaliana: Unexpected structural complexity and lack of fast responses to environmental changes (submitted to Plant Cell) [Google Scholar]
- 14.Titarenko E, Rojo E, Leon J, Sanchez-Serrano J. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiology. 1997;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]