Abstract
Cell division is a highly regulated process that has to be coordinated with cell specification and differentiation for proper development and growth of the plants. Cell cycle regulation is carried out by key proteins that control cell cycle entry, progression and exit. This regulation is controlled at different stages such as gene expression, posttranslational modification of proteins and specific proteolysis. The G1/S and the G2/M transitions are critical checkpoints of the cell cycle that are controlled, among others, by the activity of cyclin-dependent kinases (CDK). Different CDK activities, still to be fully identified, impinge on the retinoblastoma (RBR)/E2F/DP pathway as well as on the programmed proteolysis pathway. The specific degradation of proteins through the ubiquitin pathway in plants, highly controlled in time and space, is emerging as a powerful mechanism to regulate the levels and the activity of several proteins, including many cell cycle regulators.
Key Words: cell cycle, endoreplication, E2F, DP, Ubiquitin, SCF, SKP2, lateral root, Arabidopsis
Cell Cycle in Plants
The completion of the Arabidopsis and rice genomes has made possible the identification of the majority of the core cell cycle genes in plants.1–3 A significant complexity has been revealed for many key cell cycle genes, although in some cases this may have a functional redundancy.4–6 For example, there is a large collection of cyclin genes (>30 in total), grouped into four families. Several types of cyclin dependent kinases (CDK) and CDK inhibitors have been also identified.2 In addition, six E2F and two DP genes encoding transcription factors have been identified. Opposite to this diversity, only one RBR gene is present in the Arabidopsis genome.
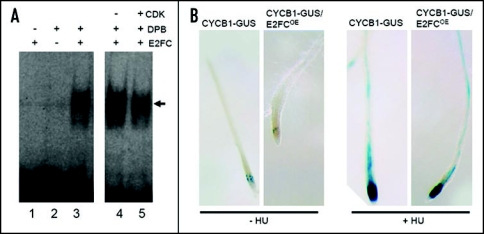
E2FA-C proteins need to heterodimerize with other protein partners, either DPA or DPB, to form an active transcription factor. E2FA and E2FB function as positive regulators of cell division, promoting cell cycle entry but also they play roles in the endocycle.7–10 However, E2FC works as negative regulator of cell division and is likely necessary for DNA endoreplication, since plants with reduced levels of E2FC showed lower levels of ploidy.11,12 E2FC forms a complex, at least, with DPB to regulate gene expression and such interaction is regulated by CDK-dependent phosphorylation.12 Thus, CDK activity reduces the binding of the heterodimer to the DNA (Fig. 1A). As a consequence, CDKA activity seems to eliminate the repressor function of E2FC-DPB. E2FC is ubiquitously expressed in the plant, with higher levels in active division zones (meristems, lateral root primordia, flower buds), in the trichomes, and the vascular tissue.11 Recent work showed that plants with reduced levels of E2FC have significantly increased levels of CYCB1;1 expression and increased number of dividing cells. On the contrary, overexpression of E2FC dramatically reduces the levels of CYCB1:1 expression. These results indicate that E2FC function as negative regulator of cell proliferation in the dividing zones.
Figure 1.
(A) CDK activity reduces binding of E2FC-DPB to DNA. EMSA assay was carried out using oligonucleotides containing the consensus E2F sites11 and recombinant proteins (GST-DPB and HIS-E2FC) produced in bacterial as described.12 Arrow points to the E2FC-DPB-DNA complexes. To test the effect of CDK-dependent phosphorylation on the DNA binding, the GST-DPB and HIS-E2FC proteins were previously incubated with CDKA-CYCA2;2 in the presence of cold ATP for 15 minutes. Afterwards, these proteins were used in the EMSA assay. CDK activity reduces the ability of E2FC-DPB to bind the E2F-DNA motif (compare lane 4 with lane 5). (B) CYCB1;1 in HU-treated E2FC overexpressor plants. CYCB1-GUS (control) or CYCB1-GUS/E2FCOE plants were grown in MS liquid medium for 5 days and then treated with 10 mM of hydroxyurea (HU) for 24 hours. Afterwards, the seedlings were stained for GUS activity as described.12 Note that the HU treatment led to a high accumulation of CYCB1;1, even when E2FC is overexpressed.
We have also found that a treatment with hydroxyurea (HU), a DNA synthesis inhibitor that induces the DNA damage response,13 increased the expression of the mitotic CYCB1;1 gene (Fig. 1B). Interestingly, we found that a treatment with HU activates the expression of CYCB1;1 even when E2FC is overexpressed. This result indicates that the E2FC-dependent repression of CYCB1;1 in the absence of DNA damage is bypassed and does not occur when the DNA damage checkpoint is activated. These preliminary data are also consistent with the complex regulation of CYCB1;1 gene expression that is being determined.14
SCFSKP2A Functions in Cell Cycle
The ubiquitin-26S proteasome pathway regulates several aspects of plant growth, development and responses to external signals (reviewed in refs. 15–17). Among them, this pathway seems to be a key regulatory mechanism that controls cell division.12,18 In a previous work, we identified two genes that encode proteins with homology to human SKP2, an F-box that regulates cell proliferation.11 Expression of SKP2A occurs in dividing zones (shoot and root meristems) and is cell cycle regulated (Jurado and del Pozo, unpublished). SKP2A forms a SCF complex that regulates the stability of both E2FC and DPB. Recent work from our laboratory showed that DPB is regulated by both phosphorylation and ubiquitin-dependent degradation, the latter involving the activity of SCFSKP2A complex.12 Similarly to E2FC, DPB accumulated in the skp2a mutant, indicating that the function of SKP2A is required for proper degradation of these transcription factors.
Lateral root primordia (LRP) are formed from founder pericycle cells, which are arrested in the G1 phase of the cell cycle.19,20 According to our data, it is conceivable that a concerted degradation of E2FC and DPB is important for cell division reactivation of these founder cells. Thus, lower levels of E2FC in RNAi-E2FC plants (e2f-r) lead to higher levels of mitotic CYCB1;1 gene expression along the root vascular bundle as well as to higher number of LRP.12 Consistent with these data, overexpression of SKP2A reduced the levels of E2FC in the roots and increased the number of LRP and (Jurado and del Pozo, unpublished). Although SKP2A seems to be required for the degradation of E2FC and DPB, it would not be surprising that the SCFSKP2A complex also regulates the stability of more cell cycle regulators. In fact, human SKP2 targets several proteins for degradation (see Ref. 12 and references herein). Keeping with this idea, it has been suggested that other cell cycle regulators in Arabidopsis, such as KRP221 or E2FB (our unpublished observations) are also degraded through the ubiquitin pathway. Whether the SCFSKP2 complexes are targeting these proteins for degradation or if this is not the case, which are the E3 responsible of such proteolysis remain to be determined.
Acknowledgements
This work has been partially supported by grants BMC2003-2131, BFU2006-5662 and BIO2004-01749 (Spanish Ministery of Science and Technology), and by an institutional grant from Fundación Ramon Areces.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/3897
References
- 1.Arabidopsis genome initiative. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature. 2002;408:816–820. doi: 10.1038/35048500. [DOI] [PubMed] [Google Scholar]
- 2.Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 4.Dewitte W, Murray JA. The plant cell cycle. Annu Rev Plant Physiol Plant Mol Biol. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez C. Coupling cell proliferation and development in plants. Nat Cell Biol. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- 6.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Gene. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 7.De Veylder L, Beeckman T, Beemster GT, de Almeida-Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inze D. Fa-DPa transcription factor. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L. The role of the Arabidopsis E2FB transcription factor in regulating Auxin-dependent cell division. Plant Cell. 2005;17:2527–2541. doi: 10.1105/tpc.105.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sozzani R, Maggio C, Varotto S, Canova S, Bergonioux C, Albani D, Cella R. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 2006;140:1355–1366. doi: 10.1104/pp.106.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Pozo JC, Boniotti MB, Gutierrez C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(SKP2) pathway in response to light. Plant Cell. 2002;14:3057–3071. doi: 10.1105/tpc.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Pozo C, Diaz-Trivino S, Cisneros N, Gutierrez C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway. Plant Cell. 2006;18:2224–2235. doi: 10.1105/tpc.105.039651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culligan K, Tissier A, Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16:1091–1104. doi: 10.1105/tpc.018903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon J, Parry G, Estelle M. The ubiquitin-proteasome pathway and plant development. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 17.Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle. Angew Chem Int Ed Engl. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- 19.Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. Dissecting Arabidopsis lateral root development. Trends Plant Sci. 2003;8:165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 20.Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inze D, Beeckman T. Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA. 2004;101:5146–5151. doi: 10.1073/pnas.0308702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verkest A, Manes C, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]