Abstract
The biological role of genomic imprinting in adult tissue is central to the consideration of transplanting uniparental embryonic stem (ES) cell-derived tissues. We have recently shown that both maternal (parthenogenetic/gynogenetic) and paternal (androgenetic) uniparental ES cells can differentiate, both in vivo in chimeras and in vitro, into adult-repopulating hematopoietic stem and progenitor cells. This suggests that, at least in some tissues, the presence of two maternal or two paternal genomes does not interfere with stem cell function and tissue homeostasis in the adult. Here, we consider implications of the contribution of uniparental cells to hematopoiesis and to development of other organ systems, notably neural tissue for which consequences of genomic imprinting are associated with a known bias in development and behavioral disorders. Our findings so far indicate that there is little or no limit to the differentiation potential of uniparental ES cells outside the normal developmental paradigm. As a potentially donor MHC-matching source of tissue, uniparental transplants may provide not only a clinical resource but also a unique tool to investigate aspects of genomic imprinting in adults.
Key words: uniparental, androgenetic, chimera, transplantation, parthenogenetic, gynogenetic, hematopoietic, neural
Mammalian Uniparental Embryos: Origin and Development
Parthenogenesis, i.e. complete development of offspring from an oocyte without male genetic contribution, is obligate in several vertebrate species of the order squamata (lizards and snakes) and can occur occasionally and spontaneously in species that normally reproduce sexually, the exception being mammals. In mammals, the two parental genomes are rendered functionally non-equivalent by a process termed genomic imprinting that results in preferential expression of certain genes (“imprinted genes”) from only one parental allele.1–8 As a consequence, completion of mammalian development requires contribution from both maternal and paternal genomes. Mouse embryos with two maternal genomes (diploid parthenogenetic (PG) embryos, or gynogenetic (GG) embryos with two maternal genomes from different oocytes; Fig. 1A) fail early post implantation due to poor development of extraembryonic tissues, and rarely develop as far as later (20–25) somite stages.1,2,9,10 Murine androgenetic (AG) embryos with two sperm-derived genomes fail even earlier, with limited development of the embryo proper but often well-developed trophoblast, and reach at most early somite stages.2,11,12 Parthenogenetic development in other mammalian species, including sheep,13,14 cattle,15 pig16,17 and rabbit18 can vary depending on the procedure used for activation of the oocyte, but does not proceed past early fetal stages, and developmental failure appears to be consistent with the stage when the conceptus begins to depend on the placenta. In species with delayed implantation, for example sheep and pig, PG fetuses can differentiate substantially and undergo organogenesis.13,16,17 Phenotypic similarities of PG conceptuses between species include growth retardation of the fetus (mouse, sheep, pig and rabbit, with the exception of sheep GG conceptuses19), suggesting that mechanisms of genomic imprinting and effects of abnormal levels of imprinted gene products can be similar between mammalian species.14 Some differences have been observed in respect to extraembryonic lineage development of maternally-derived conceptuses, which is severely hypotrophic in the mouse,1 but not in sheep19 or in rabbit.18 AG development has been investigated in sheep and bovine, indicating similar limited postimplantation viability.19,20 Human uniparental gestations are frequent and can develop into ovarian teratomas resulting from the spontaneous parthenogenetic activation of an oocyte, or, when of AG origin, form complete hydatidiform moles that exhibit abundant trophoblast (trophoblastic hyperplasia) and very poor development of embryonic tissues.21,22
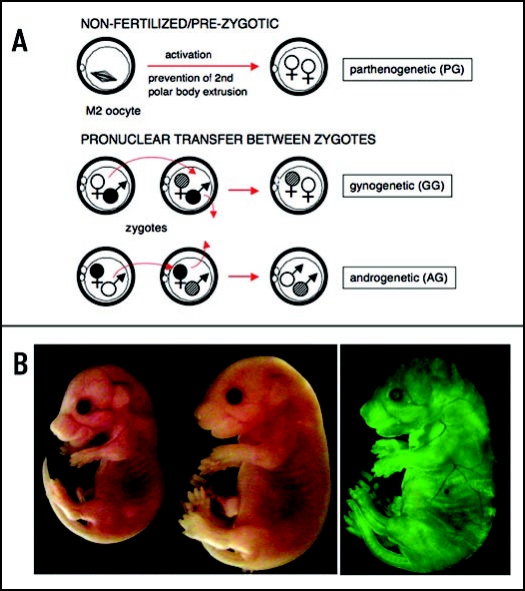
Figure 1.
Diploid uniparental embryos and overgrowth phenotype of AG ES cell chimeras. (A) Activation of an unfertilized oocyte and suppression of second polar body extrusion results in a diploid parthenogenetic embryo. GG and AG embryos with two maternal or paternal genomes from two oocytes or sperm, respectively, can be produced experimentally by pronuclear transfer between zygotes.98 Alternatively, AG embryos can be produced by in vitro fertilization99 or, presumably, by intracytoplasmic sperm injection of enucleated donor oocytes, procedures not involving fertilized embryos. (B) Overgrowth phenotype of an AG ES cell chimera (right) at E 16.5 compared to normal littermate (left). Contribution of AG cells visualized by EGFP fluorescence in E 16.5 AG chimera (AG ES cells are EGFP transgenic).
Derivation of Embryonic Stem Cells From Uniparental Embryos
Although unable to develop normally after implantation, diploid uniparental embryos can form blastocysts; and in species in which the derivation of embryonic stem (ES) cells has been established, uniparental ES cells can be derived, such as in mouse (PG23 and AG24), non-human primate (PG25), and human (PG26–28). In particular, the derivation of PG ES cells in non-human primates25 with their differentiation potential generated interest in PG ES cells as a source of patient derived tissue for transplantation.
The discussion of uniparental ES cells has mostly focused on potential advantages in respect to ethical issues and in respect to immune compatibility. Using uniparental embryos to generate ES cells subjectively side-steps some of the ethical issues associated with the destruction of potentially viable fertilized and somatic cell nuclear transfer embryos.29–31 Uniparental ES cells can contain a full MHC complement of the gamete donor.26,32 More critical to any therapeutic application, however, is the question of whether or not uniparental ES cells, derived from embryos that are severely compromised in their ability to develop due to their abnormal state of genomic imprinting, can form tissues that are functional and safe after transplantation. Genomic imprinting appears not be relevant for the formation of ES cells, but abnormal expression of imprinted genes influences the differentiation potential of ES cells.33,34 While uniparental ES cells have been termed pluripotent due to their contribution to various tissues upon blastocyst injection,24,35,36 their contribution in chimeras causes abnormal phenotypes, particularly for AG ES cells. Attempts at the formation of entirely PG ES cell-derived fetuses by tetraploid complementation resulted in developmental failure similar to PG conceptuses.35
Developmental Potential of Uniparental Cells in Chimeras
Uniparental cleavage stage blastomeres, inner cell mass (ICM) and ES cells can contribute to development, both to fetal and postnatal stages, when combined at preimplantation stages with normal mouse embryos to form chimeras.24,37–40 Chimera analyses thus provide one approach to evaluate the developmental potential of uniparental cells, but not necessarily address their therapeutic value. In chimeras, both AG and PG/GG cells have been found to contribute to most lineages including the germline.41,42 However, AG and PG/GG cells exhibit bias in their contribution to certain tissues, for example, PG cells are rarely found in skeletal muscle, whereas AG cells are frequently found in mesodermal derivatives and less in brain, indicating differences in the developmental competence of uniparental maternal and paternal cells.24,39,43–46 For both types of uniparental cells, widespread and high percentage contribution to chimeras is observed in the first half of gestation, followed by an overall reduction of contribution in the second half of gestation,39 which has been interpreted as a selective elimination process,45 however, the precise reasons for this reduction remain unclear. Consistent with the developmental potential of both types of uniparental conceptuses on their own, the phenotype of AG chimeras is more compromised than that of PG chimeras. Unlike parthenogenetic cells, AG cells cause severe defects and frequent lethality when combined with normal cells in chimeras (Fig. 1B).24,39,40 Typically, an overall contribution of more than 10% of AG cells results in embryonic and perinatal lethality.42 The propensity of AG-derived fibroblasts to undergo transformation in vitro47 is reminiscent of the formation of choriacarcinomas from naturally occuring AG conceptuses. In contrast, proliferative defects have been observed for PG cells.44,47–49 Although various properties, particularly the phenotype of ES cell chimeras generated with AG ES cells24,35,50 indicate that genomic imprinting is similar in uniparental ES cells compared to primary embryonic cells such as those derived from cleavage stage embryos or the ICM of uniparental blastocysts, some phenotypic differences exist between PG ES cell chimeras and PG aggregation chimeras.35,47 PG ES cell chimeras do not exhibit growth deficits observed with PG aggregation chimeras,35 and the exclusion of PG cells to liver or muscle,44,45 has not been similarly observed with PG ES cells,36 indicating differences in the developmental potential of these cells that could be associated with epigenetic changes resulting from ES cell derivation and culture.51–53
Potential of Uniparental Cells to Form Adult Tissues
The reasons for the limited contribution of uniparental cells in chimeras overall and the bias in contribution to some tissue types are unclear. Exclusion or bias could be explained by selection or differentiation defects of the uniparental cells during development, when abnormal levels of imprinted gene products, for instance those involved in fetal growth regulation54,55 are particularly consequential. Alternatively, uniparental cells could be intrinsically limited in their differentiation into particular cell types and would thus also not function normally in the adult. This question is particularly relevant to AG cells that cause overgrowth and organomegaly in chimeras during fetal development resulting in lethality and poor postnatal survival. Chimeras as a study tool are also limited since contribution in chimeras is stochastic, and may not reflect the full or/and intrinsic potential of cells.
One approach to investigate the consequences of uniparental origin for various tissues of a postnatal animal that also addresses their therapeutic potential, is the use of transplantation and organ regeneration paradigms, using fetal tissue or in vitro derivatives of ES cells. In several adult organs, transplanted primary fetal tissue can partially or completely replace the adult tissue through the engraftment and activity of fetal stem and progenitor cells. In rodents, such transplantation models include hematopoietic replacement (fetal liver transplant56), liver,57,58 brain59,60 and testes.61–64 Tissue transplants from midgestation stage uniparental chimeras, prior to the exclusion of uniparental cells in the second half of gestation, into adults can thus be used as an approach to bypass developmental stages during which abnormal development or exclusion of uniparental cells is frequently observed, and may permit analysis of uniparental cell functionality in adults. Uniparental ES cell-derived transplants could also be derived from in vitro differentiation strategies.
The best-studied fetal transplant paradigm is the hematopoietic reconstitution of lethally irradiated adult recipients using fetal liver hematopoietic stem cells (HSC) capable of long-term and multilineage reconstitution.65,66 This paradigm has several unique advantages to test uniparental cell potency, including that entirely donor-derived hematopoiesis can be achieved (full organ replacement from the transplant) and stringency (recipient survival depends on functionality, blood homeostasis). We have used this approach to investigate and compare the potential of murine AG and GG cells for hematopoietic differentiation in the adult.
Adult Repopulating HSCs Generated From Uniparental ES Cells
To assess the ability of uniparental ES cell derivatives to generate functional tissue-specific stem cells, we transplanted chimeric fetal liver cells from embryonic day (E) 13.5 to 14.5 AG, GG, and as a control, normal (N, derived from a fertilized embryo) ES cell chimeras into irradiated adult recipients. We then assayed contribution of EGFP transgenic AG, GG and N ES cell derivatives to the peripheral blood of recipients for more than 12 months post transplantation. Uniparental ES cell derivatives, both AG and GG cells, contributed at high levels to the hematopoietic system of recipients, comparable to N-derived cells, with no differences between N, AG or GG cells in the formation of myeloid/erythroid and lymphoid blood lineages. Repopulated recipients had a normal lifespan and no pathologies associated with uniparental cell transplants. Successful hematopoietic reconstitution of secondary recipients demonstrated the presence of self-renewing HSCs of uniparental origin (Fig. 2A).67 To exclude the possibility that cell fusion between uniparental (AG, GG) ES cell- and normal blastocyst-derived cells accounted for the ability of uniparental cells to reconstitute the hematopoietic system of recipients, we identified the origin of chimeric fetal liver transplants and adult blood cells of recipients by analysis of migratory isoforms of glucose phosphate isomerase 1 (Gpi1). ES-, blastocyst- and host-derived cells express distinct isoforms of Gpi1, such that products of cell fusion either in the fetal liver or in the adult recipient would be apparent from the presence and distribution of certain Gpi1 dimers. Using this approach, we found no evidence for cell fusion (Fig. 2B). Thus, cells with exclusively maternal or paternal genomes can form long-term and multilineage repopulating HSCs.
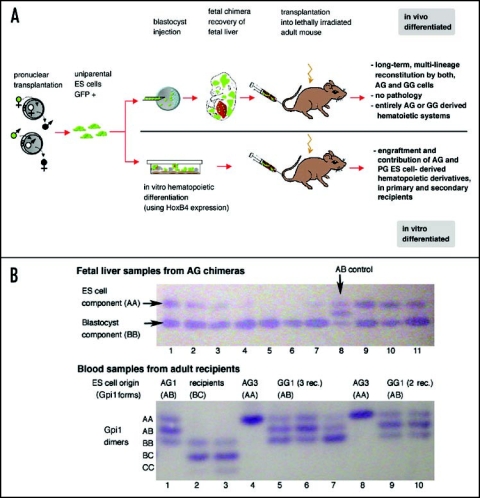
Figure 2.
Hematopoietic reconstitution by uniparental cells. (A) Outline and summary of results. (B) Glucosephosphate isomerase 1 (Gpi1) isoform analysis shows that uniparental ES-cell derived cells do not fuse with blastocyst-or host-derived cells. Gpi1 forms homo- and heterodimers. Cells homozygous for either a or b alleles contain only the respective homodimer (AA or BB; top panel, lane 1), while cells heterozygous for a and b alleles contain AA, AB and BB dimers (ES cells = AB control; top panel, lane 8). Top panel: Fetal liver samples from 10 chimeras of AG ES line 3 exhibit both ES (AA) and blastocyst (BB) components in various ratios (lanes 1–7; 9–11), but not hybrid cells that would be identified by the AB dimer. Lower panel: Peripheral blood samples of recipients (heterozygous for b and c alleles, lanes 2 and 3) reconstituted with fetal liver chimeric for AG3 (AA) contain only ES-derived cells (AA; lanes 4 and 8) but not hybrid cells, as AB dimer is not detected. Lanes 1,6,9,10: Blood of recipients of AG1 or GG1-derived cells (ES cells: AB), with a ratio of AA, AB and BB isoforms similar to that ES cells (see lane 8 upper panel), indicating entirely ES-derived blood. Lanes 5 and 7 show recipients with both ES and blastocyst-derived blood, indicated by a stronger BB band compared to AA and AB.
For any therapeutic application, transplantable cells need to be derived entirely in vitro. We ectopically expressed the homeodomain protein HoxB4 in differentiating uniparental ES cell cultures in order to obtain repopulating HSCs from ES cells. This experimental strategy has been previously successfully used to promote the in vitro formation of HSCs from ES cell cultures that exhibit multilineage and longterm engraftment in transplanted recipients.68 We established that AG and GG/PG ES cells differentiate in vitro into HSCs that engraft irradiated recipients, indicating that uniparental ES cells have the intrinsic capacity for hematopoietic differentiation (Fig. 2A).67
From chimera studies some evidence had evolved on the capacity of uniparental cells for hematopoiesis. Contribution of uniparental cells to the peripheral blood of postnatal chimeras, which for a longer period requires the activity of functional HSCs, has been described for postnatal AG and PG ES cell chimeras24,36 and, more controversially, for PG aggregation chimeras, where contribution45 as well as absence44 have been observed. In the only human PG chimera reported to date, contribution of PG cells to peripheral blood was observed to an age of more than three years, indicating PG origin of HSC.69
In postnatal chimeras produced with the same AG and GG ES cell lines used for fetal liver transplants,67 AG and GG cells contributed to the peripheral blood (Fig. 3). Although substantial and constant (n = 2; 40 and 45% over 10 and 40 weeks postnatal age), contribution of AG ES cells to the blood of postnatal chimeras was lower than in recipients of AG ES cell chimeric fetal liver transplants, whereas the GG cell proportion in blood of postnatal chimeras could be as high as in recipients of GG chimeric fetal liver transplants. Our preliminary observations indicate no bias in lineage formation by GG cells in postnatal chimeras, but variation in postnatal AG chimeras was observed, with proportionally lower contribution of AG cells to CD4+ lymphocytes and higher contribution to Gr-1+ granulocytes, while a similar lineage bias was not detected when AG chimeric fetal liver cells were transplanted. If to be substantiated, it remains to be determined whether this is related to the AG or ES cell origin of the cells, as bias of hematopoietic differentiation can occur in ES cell chimeras.70 When bone marrow of AG chimeric animals was transplanted into lethally irradiated recipients, AG-derived cells contributed at about the same percentage to the peripheral blood of these recipients as observed in the AG chimeric bone marrow donors (5 and 10–20% AG-derived, 80–95% blastocyst component of the chimera; Fig. 3). These results indicate that AG cells can also form HSC in the bone marrow of postnatal AG chimeras.
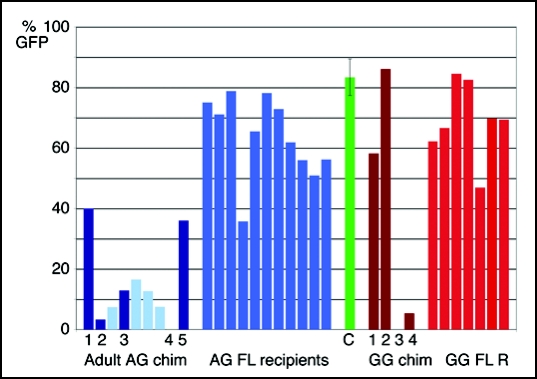
Figure 3.
Contribution of AG and GG ES cell derivatives to the blood of postnatal chimeras and recipients of fetal liver transplants. Percent of EGFP+ cells in the peripheral blood of adult AG ES cell chimeras (left, dark blue bars), recipients of bone marrow transplants from adult AG ES cell chimeras (left, light blue bars, recipients correspond to the AG chimera to their left), recipients of AG ES cell chimeric fetal liver transplants (center, bright blue), compared to EGFP transgenic animals (center, green bar, error bar indicates standard deviation (n = 17), adult GG chimeras (right, dark red bars) and recipients of GG ES cell chimeric fetal liver transplants (right, bright red bars). The EGFP transgene present in the uniparental ES cells is subject to downregulation in some blood lineages,100 such that the percentage of EGFP positive cells in the peripheral blood of transgenic control animals can vary between less than 70 and more than 90% (green bar). Therefore, evaluation of EGFP+ cells in the peripheral blood is a relative and not absolute measure for uniparental ES cell contribution.
Imprinting in Uniparental Hematopoietic Cells
The equivalence of AG and GG cells in forming repopulating HSC therefore either indicates that imprinted genes were not expressed in, or not consequential for HSC formation and differentiation, or that imprinting was relaxed. To date there is no evidence of a null mutation of a known imprinted gene resulting in a strong hematopoietic phenotype, however, the products of the imprinted Igf2 and Dlk1 genes have been associated with aspects of the regulation of HSC proliferation and differentiation,71–73 and Dlk1 null mutant mice exhibit a subtle defect in B progenitor cell frequencies.74 To ascertain how imprinting relates to the hematopoietic engraftment potential of uniparental ES cell derivatives, we have analyzed the expression and methylation status of regulatory regions of imprinted genes in uniparental-derived tissues prior and post engraftment in adult recipients, using fetal liver HSC transplants as a model. Fetal AG ES cell chimeras successfully used for hematopoietic transplants, and postnatal AG ES cell chimeras displayed imprinting-related phenotypes including overgrowth and skeletal deformities typical for AG chimeras24 consistent with phenotypic properties of AG cells. Uniparental cells isolated from chimeric fetal liver (FACS sorted for EGFP transgenic uniparental ES cell derivatives, excluding blastocyst-derived, normal cells) exhibited parent-of-origin specific expression bias for the imprinted genes Dlk-1, Igf2, Peg3 (all: preferential expression from the paternal allele) in both AG and GG cells as well as low expression levels of the maternally expressed Igf2r gene in AG cells. In contrast, no maternal-specific expression patterns were observed for p57Kip2/Cdkn1c and Meg3/Gtl2.67 However, in uniparental fetal liver-derived cells in adult recipients (uniparental CD3+ T cells; FACS sorted for EGFP expression), we did not observe expression bias, e.g. no difference between AG- and GG-derived cells, for three maternally (Ube3a, Igf2r, Meg3/Gtl2) or two paternally expressed genes (impact and U2af1-rs1), but relative expression levels for all genes except U2af1-rs1 were low.67 It is difficult to define whether these observations reflect normal basal low-level expression, tissue-specific modulation, or relaxation of the allele-specificity of expression. Allele-specific expression of imprinted genes typically does not occur in an all or none pattern, with detectable albeit lower expression from the silenced allele.75–79 We also analyzed methylation of CpG islands of three imprinting regulatory regions (the differentially methylated region (DMR) 1 of the Snrpn gene, the DMR2 of the Igf2r gene and CpG islands located the 5′ imprinting control region (ICR) of the H19 gene) in uniparental ES cells and their derivatives in adult recipients, and found considerable variation in the maintenance of imprinting marks (Fig. 4). Conservation of gametic methylation marks (H19 ICR for both AG and GG cells), partial conservation (SnrpnDMR1, conserved in AG cells and derivatives but not GG cells) and heterogeneity (Igf2rDMR2) were detected. The observation that PG and GG ES cell chimeras do not exhibit some phenotypes of PG/GG aggregation chimeras, such as growth retardation, indicate that in maternally derived uniparental ES cells, regulation of imprinted genes may be less conserved.36,47,67 Consistent with our own observations, a recent report shows re-activation of paternally expressed genes in PG ES cell lines.53 The authors observe that activation of paternally expressed genes already occurs at the blastocyst stage, indicating that extended culture may contribute to but not necessarily trigger gene expression changes. Lack of silencing of imprinted genes in uniparental fetuses suggests that for some imprinted genes, the presence of both parental genomes is required for allele-specific control of expression.80–82
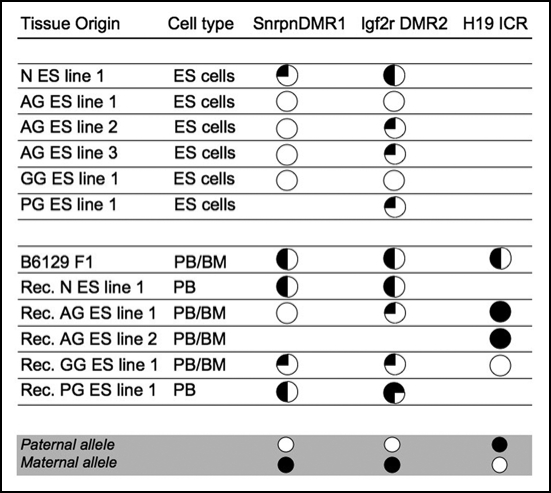
Figure 4.
Methylation of regulatory regions of imprinted genes in uniparental ES cells and their hematopoietic derivatives in recipients. Methylation analysis was performed by bisulfite sequencing.67 Samples were taken from animals with a > 95% contribution (no detectable host or blastocyst component as per Gpi1 analysis) from the ES cell derived component. Circles represent the percentage of methylation detected in all clones (full: methylated, ¼, ½ or ¾: 25, 50 or 75% methylated, empty: not methylated; approximation of methylation to nearest %). Rec., recipient; BM, bone marrow (cell type analyzed for H19); PB, peripheral blood (all other analyses for recipients). The grey bar indicates methylation patterns expected for each parental allele based on analysis of methylation in gametes.101
Although we have only examined a small proportion of known imprinted loci, our findings suggest that there is considerable flexibility or tolerance in relation to variations in imprinted gene expression and methylation in the hematopoietic system of the adult.
Neural Differentiation Capacity of Uniparental ES Cells
Consequences of genomic imprinting appear to be inconsequential for HSC formation in differentiating ES cell cultures, which could be a tissue-specific phenomenon. For the neural system, in contrast, evidence for the requirement for balanced expression of imprinted genes both during fetal development and, possibly, also in the adult exists. In chimeras, embryonic PG- and AG-derived cells exhibited substantial contribution differences to neural structures at various stages of development, suggesting that the expression of imprinted genes has consequences on cell proliferation and lineage allocation during fetal brain development. In chimeras produced by the injection of AG or GG ICM cells into normal blastocysts, the overall contribution of AG cells to brain was comparable to AG cell contribution to other tissues such as mesoderm derivatives at early midgestation stages (E12–14), but proportionally much lower in later gestation and term chimeras.39 In contrast, PG- and GG-derived cells, both in aggregation and ICM injection chimeras, appear to be sequentially eliminated from many tissues throughout gestation but are frequently found at substantial levels in the brain of later gestation and term animals.39,45,83 In the brains of E13 and, more predominantly, E17 chimeras, AG cells contributed substantially to hypothalamus, septum and preoptic area, but less than PG or control N cells to cortex and striatum, whereas PG cells accumulated in the latter and were mostly excluded from the hypothalamus.84 Contribution of PG cells furthermore resulted in hypertrophy of the telencephalon, whereas the brains of AG chimeras were smaller compared to controls and in relation to body size.
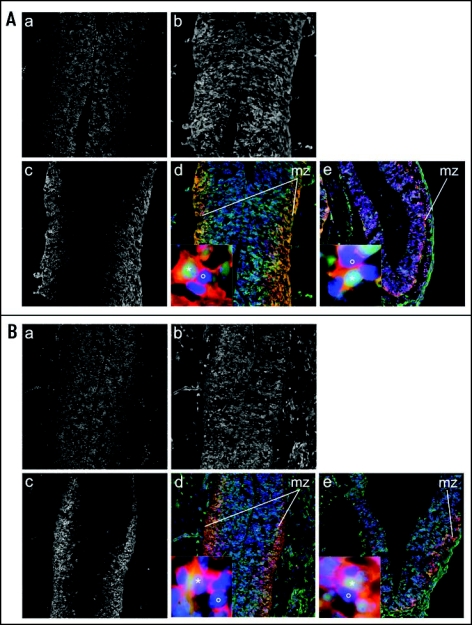
We have analyzed the contribution and differentiation of AG ES cell derivatives in midgestation (E 12.5) ES cell chimeras, and do not observe a difference in the regional distribution and neuronal differentiation of AG compared to N ES-derived cells in the diencephalon (Fig. 5).85 This result suggests, similar to observations with PG ES cells35 that developmental properties of AG ICM and ES cells can differ. It remains to be evaluated if AG ES cell derivatives exhibit similarly unrestricted contribution to the brain of chimeras at later gestational stages.
Figure 5.
Unrestricted distribution of AG ES cell derivatives in the brain of fetal chimeras. Immunostaining of brain cryosections from E 12.5 chimeras generated by blastocyst injection of ES cells. (A) AG ES cell chimera: (a) DAPI signal in transversal section including part of the diencephalon (ventral thalamus) (blue channel). (b) EGFP positive donor cells (green channel). (c) tubulin-β-III positive neurons (red channel). (d) Overlay showing EGFP positive donor cells (green), tubulin-β-III positive neurons (red) and nuclei counterstained with DAPI (blue), 100X. Insert shows a tubulin-β-III and EGFP double positive ES cell-derived cell (*) and a tubulin-β-III+ and EGFP negative blastocyst-derived cell (°) from the mantel zone (mz), 1000X. (e) Overlay and Insert of the cortex (transversal section), staining and magnification as in (d). (B) N ES chimera. Panels (a–e) are as described in panel (A).
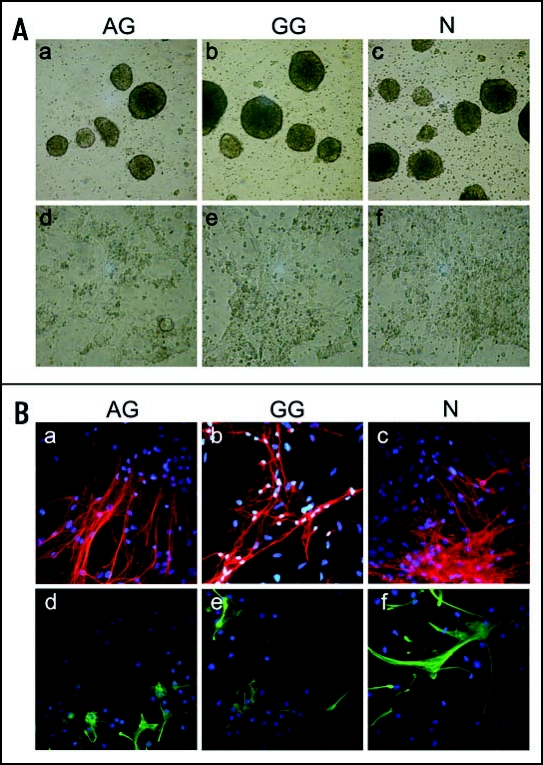
Several mental disorders including Angelmann (AS) and Prader-Willi (PWS)-Syndromes and presumably autism are associated with disturbances in the allele-specific expression of imprinted genes (both maternal and paternal alleles) during brain development.86 Consequentially, for a number of imprinted genes, developmental- and cell-type-specific imprinting has been detected in neural lineages.87 It appears that, particularly for Ube3a, an imprinted gene with maternal-specific expression in some neural lineages,88 differential imprinting is established during neurogenesis, and may depend on external cytokines.89 It is unknown whether or not these imprints would be established during neural in vitro differentiation of ES, including AG and PG/GG ES cells, limiting or permitting the formation of certain lineages. While the in vitro capacity of PG ES cells to form neural progenitors and neural progeny (dopaminergic neurons) has been shown,25 the potential of AG ES cells for neuronal differentiation is unclear. To investigate this, we cultured AG ES cells according to a multi-step protocol that facilitates ES cell differentiation towards neuronal and glial cell types90,91 and observed that AG ES cells form adherent pan-neural progenitor cells with elongated shapes, similarly to PG/GG and normal ES cells (Fig. 6A). AG pan-neural progenitors cells can further differentiate into both neuronal and glial cell types (Fig. 6B), forming β-III-tubulin positive neuronal and glial fibrillary acidic protein positive (GFAP+) astroglial cell types, with neuronal and astroglial morphology. Thus, AG ES cells have the intrinsic capacity to undergo neural commitment in vitro.85 Whether or not this is a property that distinguishes AG ES cells from AG ICM cells or not should be revealed with further analyses in respect to the ability of AG ES cells to contribute to the brains of chimeras at later gestational stages.
Figure 6.
Neural in vitro differentiation of AG, GG and N ES cells. (A) Phase contrast images of AG, GG and N ES cells at progressive stages of in vitro differentiation. (a–c) embryoid bodies derived from AG, GG and N ES cells (day 4 of differentiation), magnification: 50X. (d–f) pan-neural progenitor cells (day 12 of differentiation), 100X. (B) Immunostaining of pan-neural progenitor cell-derived neuronal and glial cells. Pan-neural progenitor cells were cultured for 13 days under conditions for neural differentiation. (a–c) tubulin-β-III positive neuronal cells (red). (d–f) GFAP+ astroglial cells (green). Nuclei were counterstained with DAPI, magnification: 200X.
Utility of Uniparental ES Cells
Therapeutic utility of uniparental ES cell derivatives depends on whether the function and balanced expression of imprinted genes is required for formation of the respective tissue or tissue stem cell, as well as normal function of the graft subsequent to transplantation. While the necessity of balanced expression of imprinted genes during development has extensively studied, for example in the regulation of fetal growth and placental development7,92,93 it is much less clear to what degree genomic imprinting is required in adult tissues.8,94 Unregulated levels of some imprinted gene products are associated with neoplasia in neonates and adults,95–97 although probably in association with dysregulation of non-imprinted genes. Thus, the tolerance for loss of imprinting or loss of heterozygosity for imprinted loci in adult tissues remains a developmentally intriguing question that needs to be explored. Investigating the ability of uniparental ES cells to recapitulate specific developmental pathways in vitro, combined with transplantation models, provides a unique tool to determine how imprinting relates to the formation of tissue specific stem and progenitor cells.
Acknowledgements
We thank Teresa Jordan for animal husbandry. This work was supported by National Institutes of Health (NIH) Grant 1 RO3 HD045291-01 (K.J.M.), by the Stem Cell Research Foundation (K.J.M.), in part, by the Marion Dilley and David George Jones Funds and the Commonwealth and General Assembly of Pennsylvania (K.J.M) and by the Deutsche Forschungsgemeinschaft DFG GRK 1048 “Molecular basis of organ development in vertebrates” (A.M.M.).
Abbreviations
- ES cell
embryonic stem cell
- PG
parthenogenetic
- AG
androgenetic
- GG
gynogenetic
- HSC
hematopoietic stem cell
- ICM
inner cell mass
Footnotes
Previously published online as an Organogenesis E-publication: www.landesbioscience.com/journals/organogenesis/article/6123
References
- 1.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 2.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 3.Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 4.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 5.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 6.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 7.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Solter D. Imprinting today: end of the beginning or beginning of the end? Cytogenet Genome Res. 2006;113:12–16. doi: 10.1159/000090809. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman MH, Barton SC, Surani MA. Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature. 1977;265:53–55. doi: 10.1038/265053a0. [DOI] [PubMed] [Google Scholar]
- 10.Surani MA, Barton SC. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science. 1983;222:1034–1036. doi: 10.1126/science.6648518. [DOI] [PubMed] [Google Scholar]
- 11.Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 12.Surani MAH, Barton SC, Norris ML. Nuclear transplantation in the mouse: Heritable differences between parental genomes after activation of the embryonic genome. Cell. 1986;45:127–136. doi: 10.1016/0092-8674(86)90544-1. [DOI] [PubMed] [Google Scholar]
- 13.Loi P, Ledda S, Fulka J, Jr, Cappai P, Moor RM. Development of parthenogenetic and cloned ovine embryos: effect of activation protocols. Biol Reprod. 1998;58:1177–1187. doi: 10.1095/biolreprod58.5.1177. [DOI] [PubMed] [Google Scholar]
- 14.Feil R, Khosla S, Cappai P, Loi P. Genomic imprinting in ruminants: allele-specific gene expression in parthenogenetic sheep. Mamm Genome. 1998;9:831–834. doi: 10.1007/s003359900876. [DOI] [PubMed] [Google Scholar]
- 15.Fukui Y, Sawai K, Furudate M, Sato N, Iwazumi Y, Ohsaki K. Parthenogenetic development of bovine oocytes treated with ethanol and cytochalasin B after in vitro maturation. Mol Reprod Dev. 1992;33:357–362. doi: 10.1002/mrd.1080330318. [DOI] [PubMed] [Google Scholar]
- 16.Kure-bayashi S, Miyake M, Okada K, Kato S. Successful implantation of in vitro-matured, electro-activated oocytes in the pig. Theriogenology. 2000;53:1105–1119. doi: 10.1016/S0093-691X(00)00256-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, King T, Dobrinsky J, Harkness L, Ferrier T, Bosma W, Schreier LL, Guthrie HD, DeSousa P, Wilmut I. In vitro and in vivo developmental competence of ovulated and in vitro matured porcine oocytes activated by electrical activation. Cloning Stem Cells. 2003;5:355–365. doi: 10.1089/153623003772032853. [DOI] [PubMed] [Google Scholar]
- 18.Ozil JP. The parthenogenetic development of rabbit oocytes after repetitive pulsatile electrical stimulation. Development. 1990;109:117–127. doi: 10.1242/dev.109.1.117. [DOI] [PubMed] [Google Scholar]
- 19.Hagemann LJ, Peterson AJ, Weilert LL, Lee RS, Tervit HR. In vitro and early in vivo development of sheep gynogenones and putative androgenones. Mol Reprod Dev. 1998;50:154–162. doi: 10.1002/(SICI)1098-2795(199806)50:2<154::AID-MRD5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 20.Lagutina I, Lazzari G, Duchi R, Galli C. Developmental potential of bovine androgenetic and parthenogenetic embryos: a comparative study. Biol Reprod. 2004;70:400–405. doi: 10.1095/biolreprod.103.021972. [DOI] [PubMed] [Google Scholar]
- 21.Mutter GL. Role of imprinting in abnormal human development. Mutat Res. 1997;396:141–147. doi: 10.1016/s0027-5107(97)00180-2. [DOI] [PubMed] [Google Scholar]
- 22.Fisher RA, Hodges MD. Genomic imprinting in gestational trophoblastic disease—a review. Placenta. 2003;24(Suppl A):S111–S118. doi: 10.1053/plac.2002.0939. [DOI] [PubMed] [Google Scholar]
- 23.Robertson EJ, Kaufman MH, Bradley A, Evans MJ. Isolation, properties, and karyotype analysis of pluripotential (EK) cell lines from normal and parthenogenetic embryos. In: Silver LM, Martin GR, Strickland S, editors. Teratocarcinomal Stem Cells Cold Spring Harbor Conferences on Cell Proliferation. New York: Cold Spring Harbor Laboratory; 1983. pp. 647–663. [Google Scholar]
- 24.Mann JR, Gadi I, Harbison ML, Abbondanzo SJ, Stewart CL. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: Implications for genetic imprinting. Cell. 1990;62:251–260. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- 25.Cibelli JB, Grant KA, Chapman KB, Cunniff K, Worst T, Green HL, Walker SJ, Gutin PH, Vilner L, Tabar V, Dominko T, Kane J, Wettstein PJ, Lanza RP, Studer L, Vrana KE, West MD. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 26.Revazova ES, Turovets NA, Kochetkova OD, Kindarova LB, Kuzmichev LN, Janus JD, Pryzhkova MV. Patient-Specific Stem Cell Lines Derived from Human Parthenogenetic Blastocysts. Cloning Stem Cells. 2007 doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 27.Revazova ES, Turovets NA, Kochetkova OD, Agapova LS, Sebastian JL, Pryzhkova MV, Smolnikova VI, Kuzmichev LN, Janus JD. HLA Homozygous Stem Cell Lines Derived from Human Parthenogenetic Blastocysts. Cloning Stem Cells. 2007 doi: 10.1089/clo.2007.0063. [DOI] [PubMed] [Google Scholar]
- 28.Mai Q, Yu Y, Li T, Wang L, Chen MJ, Huang SZ, Zhou C, Zhou Q. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2007;17:1008–1019. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- 29.Daley GQ, Ahrlund Richter L, Auerbach JM, Benvenisty N, Charo RA, Chen G, Deng HK, Goldstein LS, Hudson KL, Hyun I, Junn SC, Love J, Lee EH, McLaren A, Mummery CL, Nakatsuji N, Racowsky C, Rooke H, Rossant J, Scholer HR, Solbakk JH, Taylor P, Trounson AO, Weissman IL, Wilmut I, Yu J, Zoloth L. Ethics. The ISSCR guidelines for human embryonic stem cell research. Science. 2007;315:603–604. doi: 10.1126/science.1139337. [DOI] [PubMed] [Google Scholar]
- 30.Hipp J, Atala A. Tissue engineering, stem cells, cloning, and parthenogenesis: new paradigms for therapy. J Exp Clin Assist Reprod. 2004;1:3. doi: 10.1186/1743-1050-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaenisch R. Human cloning - the science and ethics of nuclear transplantation. N Engl J Med. 2004;351:2787–2791. doi: 10.1056/NEJMp048304. [DOI] [PubMed] [Google Scholar]
- 32.Kim K, Lerou P, Yabuuchi A, Lengerke C, Ng K, West J, Kirby A, Daly MJ, Daley GQ. Histocompatible embryonic stem cells by parthenogenesis. Science. 2007;315:482–486. doi: 10.1126/science.1133542. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 34.Morali OG, Jouneau A, McLaughlin KJ, Thiery JP, Larue L. IGF-II promotes mesoderm formation. Dev Biol. 2000;227:133–145. doi: 10.1006/dbio.2000.9875. [DOI] [PubMed] [Google Scholar]
- 35.Allen ND, Barton SC, Hilton K, Norris ML, Surani MA. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 36.Sturm KS, Berger CN, Zhou SX, Dunwoodie SL, Tan S, Tam PP. Unrestricted lineage differentiation of parthenogenetic ES cells. Dev Genes Evol. 1997;206:377–388. doi: 10.1007/s004270050067. [DOI] [PubMed] [Google Scholar]
- 37.Stevens LC, Varnum DS, Eicher EM. Viable chimaeras produced from normal and parthenogenetic mouse embryos. Nature. 1977;269:515–517. doi: 10.1038/269515a0. [DOI] [PubMed] [Google Scholar]
- 38.Surani MA, Barton SC, Kaufman MH. Development to term of chimaeras between diploid parthenogenetic and fertilised embryos. Nature. 1977;270:601–603. doi: 10.1038/270601a0. [DOI] [PubMed] [Google Scholar]
- 39.Barton SC, Ferguson-Smith AC, Fundele R, Surani MA. Influence of paternally imprinted genes on development. Development. 1991;113:679–687. doi: 10.1242/dev.113.2.679. [DOI] [PubMed] [Google Scholar]
- 40.Mann JR, Stewart CL. Development to term of mouse androgenetic aggregation chimeras. Development. 1991;113:1325–1333. doi: 10.1242/dev.113.4.1325. [DOI] [PubMed] [Google Scholar]
- 41.Stevens LC. Totipotent cells of parthenogenetic origin in a chimaeric mouse. Nature. 1978;276:266–267. doi: 10.1038/276266a0. [DOI] [PubMed] [Google Scholar]
- 42.Narasimha M, Barton SC, Surani MA. The role of the paternal genome in the development of the mouse germ line. Curr Biol. 1997;7:881–884. doi: 10.1016/s0960-9822(06)00377-0. [DOI] [PubMed] [Google Scholar]
- 43.Paldi A, Nagy A, Markkula M, Barna I, Dezso L. Postnatal development of parthenogenetic in equilibrium with fertilized mouse aggregation chimeras. Development. 1989;105:115–118. doi: 10.1242/dev.105.1.115. [DOI] [PubMed] [Google Scholar]
- 44.Nagy A, Sass M, Markkula M. Systematic non-uniform distribution of parthenogenetic cells in adult mouse chimaeras. Development. 1989;106:321–324. doi: 10.1242/dev.106.2.321. [DOI] [PubMed] [Google Scholar]
- 45.Fundele R, Norris ML, Barton SC, Reik W, Surani MA. Systematic elimination of parthenogenetic cells in mouse chimeras. Development. 1989;106:29–35. doi: 10.1242/dev.106.1.29. [DOI] [PubMed] [Google Scholar]
- 46.Fundele RH, Norris ML, Barton SC, Fehlau M, Howlett SK, Mills WE, Surani MA. Temporal and spatial selection against parthenogenetic cells during development of fetal chimeras. Development. 1990;108:203–211. doi: 10.1242/dev.108.1.203. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez L, Kozlov S, Piras G, Stewart CL. Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc Natl Acad Sci U S A. 2003;100:13344–13349. doi: 10.1073/pnas.2234026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jagerbauer EM, Fraser A, Herbst EW, Kothary R, Fundele R. Parthenogenetic stem cells in postnatal mouse chimeras. Development. 1992;116:95–102. doi: 10.1242/dev.116.1.95. [DOI] [PubMed] [Google Scholar]
- 49.Newman-Smith ED, Werb Z. Stem cell defects in parthenogenetic peri-implantation embryos. Development. 1995;121:2069–2077. doi: 10.1242/dev.121.7.2069. [DOI] [PubMed] [Google Scholar]
- 50.Mann JR. Properties of androgenetic and parthenogenetic mouse embryonic stem cell lines; are genetic imprints conserved? Seminars Dev Biol. 1992;3:77–85. [Google Scholar]
- 51.Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R. Altered imprinted gene methylation and expression in completely ES cell- derived mouse fetuses: association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 52.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 53.Jiang H, Sun B, Wang W, Zhang Z, Gao F, Shi G, Cui B, Kong X, He Z, Ding X, Kuang Y, Fei J, Sun YJ, Feng Y, Jin Y. Activation of paternally expressed imprinted genes in newly derived germline-competent mouse parthenogenetic embryonic stem cell lines. Cell Res. 2007;17:792–803. doi: 10.1038/cr.2007.70. [DOI] [PubMed] [Google Scholar]
- 54.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaughlin KJ, Kochanowski H, Solter D, Schwarzkopf G, Szabo PE, Mann JR. Roles of the imprinted gene Igf2 and paternal duplication of distal chromosome 7 in the perinatal abnormalities of androgenetic mouse chimeras. Development. 1997;124:4897–4904. doi: 10.1242/dev.124.23.4897. [DOI] [PubMed] [Google Scholar]
- 56.Fleischman RA, Custer RP, Mintz B. Totipotent hematopoietic stem cells: normal self-renewal and differentiation after transplantation between mouse fetuses. Cell. 1982;30:351–359. doi: 10.1016/0092-8674(82)90233-1. [DOI] [PubMed] [Google Scholar]
- 57.Dabeva MD, Petkov PM, Sandhu J, Oren R, Laconi E, Hurston E, Shafritz DA. Proliferation and differentiation of fetal liver epithelial progenitor cells after transplantation into adult rat liver. Am J Pathol. 2000;156:2017–2031. doi: 10.1016/S0002-9440(10)65074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nierhoff D, Ogawa A, Oertel M, Chen YQ, Shafritz DA. Purification and characterization of mouse fetal liver epithelial cells with high in vivo repopulation capacity. Hepatology. 2005;42:130–139. doi: 10.1002/hep.20735. [DOI] [PubMed] [Google Scholar]
- 59.Sayles M, Jain M, Barker RA. The cellular repair of the brain in Parkinson's disease—past, present and future. Transpl Immunol. 2004;12:321–342. doi: 10.1016/j.trim.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 60.Nakao N, Itakura T. Fetal tissue transplants in animal models of Huntington's disease: the effects on damaged neuronal circuitry and behavioral deficits. Prog Neurobiol. 2000;61:313–338. doi: 10.1016/s0301-0082(99)00058-1. [DOI] [PubMed] [Google Scholar]
- 61.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang FX, Short RV. Different fate of primordial germ cells and gonocytes following transplantation. Apmis. 1998;106:58–62. doi: 10.1111/j.1699-0463.1998.tb01319.x. discussion -3. [DOI] [PubMed] [Google Scholar]
- 63.Ohta H, Wakayama T, Nishimune Y. Commitment of fetal male germ cells to spermatogonial stem cells during mouse embryonic development. Biol Reprod. 2004;70:1286–1291. doi: 10.1095/biolreprod.103.024612. [DOI] [PubMed] [Google Scholar]
- 64.Chuma S, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Hosokawa M, Nakatsuji N, Ogura A, Shinohara T. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development. 2005;132:117–122. doi: 10.1242/dev.01555. [DOI] [PubMed] [Google Scholar]
- 65.Jordan CT, McKearn JP, Lemischka IR. Cellular and developmental properties of fetal hematopoietic stem cells. Cell. 1990;61:953–963. doi: 10.1016/0092-8674(90)90061-i. [DOI] [PubMed] [Google Scholar]
- 66.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckardt S, Leu NA, Bradley HL, Kato H, Bunting KD, Mclaughlin KJ. Hematopoietic reconstitution with androgenetic and gynogenetic stem cells. Genes & Development. 2007;21:409–419. doi: 10.1101/gad.1524207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 69.Strain L, Warner JP, Johnston T, Bonthron DT. A human parthenogenetic chimaera. Nat Genet. 1995;11:164–169. doi: 10.1038/ng1095-164. [DOI] [PubMed] [Google Scholar]
- 70.Berger CN, Tam PP, Sturm KS. The development of haematopoietic cells is biased in embryonic stem cell chimaeras. Dev Biol. 1995;170:651–663. doi: 10.1006/dbio.1995.1244. [DOI] [PubMed] [Google Scholar]
- 71.Zhang CC, Lodish HF. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- 72.Laborda J. The role of the epidermal growth factor-like protein dlk in cell differentiation. Histol Histopathol. 2000;15:119–129. doi: 10.14670/HH-15.119. [DOI] [PubMed] [Google Scholar]
- 73.Visan I, Yuan JS, Tan JB, Cretegny K, Guidos CJ. Regulation of intrathymic T-cell development by Lunatic Fringe- Notch1 interactions. Immunol Rev. 2006;209:76–94. doi: 10.1111/j.0105-2896.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 74.Sakajiri S, O'Kelly J, Yin D, Miller CW, Hofmann WK, Oshimi K, Shih LY, Kim KH, Sul HS, Jensen CH, Teisner B, Kawamata N, Koeffler HP. Dlk1 in normal and abnormal hematopoiesis. Leukemia. 2005;19:1404–1410. doi: 10.1038/sj.leu.2403832. [DOI] [PubMed] [Google Scholar]
- 75.Szabo P, Mann JR. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development. 1994;120:1651–1660. doi: 10.1242/dev.120.6.1651. [DOI] [PubMed] [Google Scholar]
- 76.Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 77.Szabo PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 78.Matsuoka S, Thompson JS, Edwards MC, Bartletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc Natl Acad Sci U S A. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burns JL, Jackson DA, Hassan AB. A view through the clouds of imprinting. Faseb J. 2001;15:1694–1703. doi: 10.1096/fj.010085rev. [DOI] [PubMed] [Google Scholar]
- 80.Sotomaru Y, Kawase Y, Ueda T, Obata Y, Suzuki H, Domeki I, Hatada I, Kono T. Disruption of imprinted expression of U2afbp-rs/U2af1-rs1 gene in mouse parthenogenetic fetuses. J Biol Chem. 2001;276:26694–26698. doi: 10.1074/jbc.M101367200. [DOI] [PubMed] [Google Scholar]
- 81.Sotomaru Y, Katsuzawa Y, Hatada I, Obata Y, Sasaki H, Kono T. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J Biol Chem. 2002;277:12474–12478. doi: 10.1074/jbc.M109212200. [DOI] [PubMed] [Google Scholar]
- 82.Ogawa H, Wu Q, Komiyama J, Obata Y, Kono T. Disruption of parental-specific expression of imprinted genes in uniparental fetuses. FEBS Lett. 2006;580:5377–5384. doi: 10.1016/j.febslet.2006.08.087. [DOI] [PubMed] [Google Scholar]
- 83.Bender R, Surani MA, Kothary R, Li L, Furst DO, Christ B, Fundele R. Tissue specific loss of proliferative capacity of parthenogenetic cells in fetal mouse chimeras. Roux Arch Devel Biol. 1995;204:436–443. doi: 10.1007/BF00360851. [DOI] [PubMed] [Google Scholar]
- 84.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 85.Dinger TC, Eckardt S, Choi SW, Camarero G, Kurosaka S, Hornich V, McLaughlin KJ, Mueller AM. Androgenetic embryonic stem cells form neural progenitor cells in vivo and in vitro. Stem Cells. 2008 doi: 10.1634/stemcells.2007-0877. In press. [DOI] [PubMed] [Google Scholar]
- 86.Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell Mol Life Sci. 2007;64:947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kishino T. Imprinting in neurons. Cytogenet Genome Res. 2006;113:209–214. doi: 10.1159/000090834. [DOI] [PubMed] [Google Scholar]
- 88.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 89.Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- 90.Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- 91.Brustle O, Spiro AC, Karram K, Choudhary K, Okabe S, McKay RD. In vitro-generated neural precursors participate in mammalian brain development. Proc Natl Acad Sci U S A. 1997;94:14809–14814. doi: 10.1073/pnas.94.26.14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 93.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65(Suppl 3):50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- 94.Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tycko B. Genomic imprinting and cancer. Results Probl Cell Differ. 1999;25:133–169. doi: 10.1007/978-3-540-69111-2_7. [DOI] [PubMed] [Google Scholar]
- 96.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 97.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- 98.McGrath J, Solter D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion. Science. 1983;220:1300–1302. doi: 10.1126/science.6857250. [DOI] [PubMed] [Google Scholar]
- 99.Kono T, Sotomaru Y, Sato Y, Nakahara T. Development of androgenetic mouse embryos produced by in vitro fertilization of enucleated oocytes. Mol Reprod Dev. 1993;34:43–46. doi: 10.1002/mrd.1080340107. [DOI] [PubMed] [Google Scholar]
- 100.Anderson DA, Wu Y, Jiang S, Zhang X, Streeter PR, Spangrude GJ, Archer DR, Fleming WH. Donor marker infidelity in transgenic hematopoietic stem cells. Stem Cells. 2005;23:638–643. doi: 10.1634/stemcells.2004-0325. [DOI] [PubMed] [Google Scholar]
- 101.Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]