Abstract
Brassinosteroids (BRs) are steroid phytohormones required for plant growth and development. The perception of BRs at the plasma membrane initiates intracellular signaling and induces dephosphorylation of two key transcription factors, BZR1 and BZR2/BES1. Phosphorylation of these factors is modulated by the GSK3 kinase BIN2 and phosphatase BSU1 and, in turn, controls DNA binding, protein stabilization, or/and nuclear translocation of BZR1 and BZR2/BES1. However, cytosolic signaling events and the biological roles of phosphorylation in BR signaling are still largely unknown. Recently, we demonstrated that BZR1 itself acts as a cytosolic signaling mediator and regulates expression of BR-responsive genes via phosphorylation-mediated nucleocytoplasmic shuttling. BIN2-mediated phosphorylation mediates nuclear export of BZR1 via interaction with a 14-3-3 protein, while BR activated phosphatases induce nuclear import of BZR1. The temporal and spatial expression of BIN2 appears to be essential in BR signaling. In this addendum, we summarize new findings in BR signaling and discuss the possibility that light and brassinosteroid signals intersect at BIN2 expression.
Key words: brassinosteroid signaling, BZR1, 14-3-3 proteins, Arabidopsis thaliana, nucleocytoplasmic shuttling, photomorphogenesis
Brassinosteroids (BRs), a group of steroid phytohormones, are essential for plant growth and for developmental processes such as seed germination, stem elongation, vascular differentiation, photomorphogenesis, and stress responses.1–4 BR initiates its signaling cascade through a plasma membrane-associated receptor kinase complex composed of BRI1 (BRaSSInoSteRoId InSenSItIVe 1) and BaK1 (BRI1 aSSoCIated ReCePtoR KInaSe1).5–7 BR signals perceived at the membrane are transmitted to the nucleus and activate at least two key transcriptional regulators, BZR1 (BRASSINAZOLE RESISTANT1) and BZR2/ BES1 (BRASSINAZOLE RESISTANT2/ BRI1-EMS-SUPPRESSOR 1) via dephosphorylation.8,9 The phosphorylation status of the two transcriptional regulators is tightly regulated by the activity of the BIN2 kinase and BSU1 (BRI1 SUPPRESSOR1) phosphatase, and is currently regarded as the most reliable readout in BR signaling.10–12 However, lack of accurate information on the mechanism of action and subcellular localization of each signaling component hinders understanding of the precise biological roles of phosphorylation events in BR signaling.
Phosphorylation of BZR1 and BZR2/BES1 may be involved in the protein stabilization, nuclear accumulation of these two factors, as well as their interaction with targeted dna cis-elements. In a recent issue of Plant Cell,13 we clearly showed that phosphorylation of BZR1 plays a key role in dynamic shuttling of BZR1 between the cytoplasm and the nucleus. BR-activated phosphatases dephosphorylate BZR1 and induce rapid nuclear localization. In the absence of BR signaling, BIN2-mediated phosphorylation events increase nuclear export of BZR1 through an interaction with 14-3-3 proteins. In this study, we identified two functional domains including 10 serine/threonine residues together which have regulatory roles in BIN2-mediated phosphorylation and nuclear export of BZR1. The first domain, positioned between BZR1 S110 and S138, appears to contain the primary BIN2 phosphorylation sites of BZR1. Interestingly, the second domain, which contains a conserved motif for interacting with 14-3-3 proteins, is critical for BIN2-mediated nuclear export of BZR1. Other recent studies of BZR1 and its rice homolog OsBZR1 also suggest that 14-3-3 proteins play a central role in cytoplasmic retention of BZR1, which is likely due to increased nuclear export mediated by BZR1 phosphorylation.14,15 In the current study, mutations of putative phosphorylation sites and the 14-3-3 recognition site led to constitutive nuclear localization of BZR1 in transgenic Arabidopsis and conferred strong BR-response phenotypes including brassinazole insensitivity, curled leaves, and facilitated hypocotyl elongation. BZR1 and BZR2/BES1 share an 88% amino acid sequence identity and conservation of the two putative regulatory domains identified in this study, suggesting that they may be similarly regulated. We have experimentally confirmed that the signaling output of BZR2/BES1 is also regulated by nucleocytoplasmic shuttling in a tissue and developmental stage specific manner (data not shown).
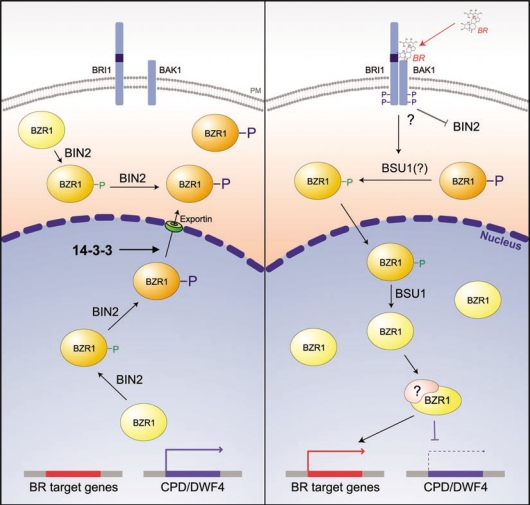
Based on our study, we have proposed a new model for the BR signal transduction pathway (Fig. 1). In the resting state, or in tissues that are not receiving BR signals, BIN2-mediated phosphorylation mediates the cytoplasmic localization of BZR1, minimizing positive BR signaling output in cells. Nuclear BIN2-mediated phosphorylation not only regulates the DNA-binding affinity of BZR1 for its target genes, but also induces export of BZR1 from the nucleus to prevent BR signaling. 14-3-3 proteins specifically interact with phosphorylated BZR1 and act as molecular chaperones to mediate the nuclear export and cytoplasmic retention of BZR1. In the BR-stimulated growth state, hyper-phosphorylated BZR1 proteins that have accumulated in the cytoplasm are targets of yet unidentified cytoplasmic phosphatases and, subsequently, the dephosphorylated proteins are imported into the nucleus. Interestingly, we observed that BSU1, a kelch-repeat phosphatase, facilitates dephosphorylation and nuclear translocation of BZR1.13,15 Although Mora-Garcia et al reported that BSU1 is constitutively localized in the nucleus,12 we found that BSU1 localizes to both the nucleus and cytoplasm in protoplasts (data not shown). Detailed examination of BSU1 localization in different tissues and developmental stages will be important to fully understand its roles in BR signaling, particularly in regard to the spatial distribution of BZR1. Physiological analysis of BZR1S130/134A, BZR1S173A, and BZR1T177A transgenic plants has suggested that the subcellular distribution of BZR1 is a primary mechanism regulating the expression of target genes in response to BR signaling.13 This mechanism may allow plants to rapidly respond to environmental or developmental stimuli without de novo synthesis of these important transcription factors. In addition, our study also strongly suggested that BZR1 itself acts as a cytosolic signaling mediator to transmit BR signals to the nucleus.
Figure 1.
A model of brassinosteroid signaling in Arabidopsis. Without a BR signal, BZR1 is hyper-phosphorylated by BIN2 kinase, interacts with a 14-3-3 protein, and is then translocated from the nucleus into the cytoplasm via an unknown exportin. Upon binding of the BR ligand, the BR signal initiated from the BRI1/BAK1 receptor complex inactivates BIN2, and/or activates the cytoplasmic-nuclear phosphatase BSU1, leading to dephosphorylation of BZR1 and its translocation into the nucleus. The hypo-phosphorylated BZR1 activates transcription of various BR target genes and also suppresses expression of CPD and DWF4, which could be synergistically regulated by another unknown factor, perhaps BIM1.21
As hypothesized from our work, the opposing actions of BIN2 and BSU1 could act as a BR-activated toggle switch converting the resting state to the BR-stimulated growth state. The temporal and spatial regulation of BIN2 or BSU1 expression may determine the magnitude of BR responses by modulating the amount of BZR1 and BZR2/BES1 proteins in the nucleus. In contrast to ubiquitously expressed BSU1,12 BIN2 expression appears to be tightly regulated by intrinsic developmental programs or environmental signals such as exposure to light and hormones.16 We found that BIN2 expression is highly correlated with auxin homeostasis during lateral root development (data not shown). A gain-of-function mutant of BIN2, ucu1, also exhibited alterations in sensitivity to auxin and BR, indicating that BIN2 probably plays an important role in cross-talk between multiple signaling cascades during plant developmental processes. In addition, the expression pattern of BIN2 in the elongation region of hypocotyls, petioles, and roots differs according to light exposure. For example, we could not detect BIN2 expression in dark-grown hypocotyls, but only in light-grown hypocotyls (data not shown). Light signals control BR homeostasis by regulating expression of BR biosynthesis genes such as Pra2 and CPD.17,18 BIN2 negatively regulates BR-mediated skotomorphogenic development,10,11 but induces expression of CPD and DWF4 by suppressing the activity of BeS1 and BZR1.19,20 It is possible that light-dependent temporal and spatial expression of BIN2 plays a critical role in photomorphogenesis by regulating the BR signaling pathway and/or BR homeostasis.
Acknowledgement
This work was supported by grants from the Plant Diversity Research Center and the KOSEF (M10601000194-06N0100-194 10) of the MOST, the Plant Signaling Network Research Center, and the Korea Research Foundation (KRF-2005-070-C00129, R08-2003-000-10819-0) of the MOEHRD.
Footnotes
Previously published online as a Plant Signaling & behavior E-publication: www.landesbioscience.com/journals/psb/article/5240
References
- 1.Cano-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 2.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 5.He Z, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 6.Li J, et al. BaK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 7.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 10.Choe S, et al. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai MY, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dornelas MC, Wittich P, von Recklinghausen I, van Lammeren A, Kreis M. Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multi-gene family. Plant Mol Biol. 1999;39:137–147. doi: 10.1023/a:1006102812280. [DOI] [PubMed] [Google Scholar]
- 17.Bancos S, et al. Diurnal regulation of the brassinosteroid-biosynthetic CPD gene in Arabidopsis. Plant Physiol. 2006;141:299–309. doi: 10.1104/pp.106.079145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang JG, et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105:625–636. doi: 10.1016/s0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- 19.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 21.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]