Abstract
MicroRNAs (miRNAs) 165 and 166 are able to cleave their target mRNAs of HD-ZIP III genes, thus regulating the functions of these genes. Although it is generally accepted that both miR165 and miR166 perform the same functions in the regulation of HD-ZIP III genes in Arabidopsis, no experimental data are available to support this notion. Recent work has shown that overexpression of miR166 downregulates the expression of three HD-ZIP III genes, ATHB-9/PHV, ATHB-14/PHB and ATHB-15, which in turn recapitulates the phenotypes of simultaneous loss-of-function mutations of these genes. In the March issue of Plant & Cell Physiology, we have demonstrated that overexpression of miR165 leads to the down-regulation of all five HD-ZIP III genes, and concomitantly recapitulates the phenotypes of loss-of-function mutation of IFL1/REV and those of simultaneous loss-of-function mutations of IFL1/REV, ATHB-9/PHV and ATHB-14/PHB. These results indicate that miR165 and miR166 differentially regulate the functions of HD-ZIP III genes in Arabidopsis. In this addendum, we show that overexpression of the antisense form of the miR165a gene leads to formation of amphivasal vascular bundles, a phenotype reminiscent of that of the dominant mutation of IFL1/REV. This finding provides direct evidence for a role of miR165 in regulation of vascular patterning.
Key Words: HD-ZIP III genes, miR165, miR166, organ polarity, vascular patterning
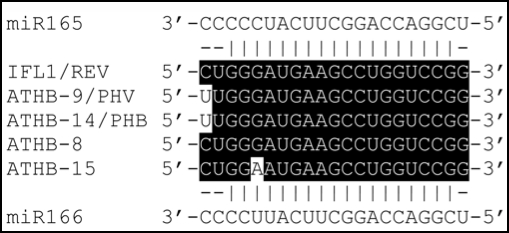
miRNAs have been shown to play important roles in diverse processes in plants, such as growth and development, and stress and disease responses.1–4 They regulate the functions of their target genes by binding to the complementary sequences in the target transcripts and subsequently causing translational attenuation or transcript cleavage. In the Arabidopsis genome, a number of miRNAs have been identified by computational prediction and sequencing of small RNAs.4 Among them, two miRNAs, miR165 and miR166, differ in only one nucleotide in their mature RNA sequences (Fig. 1).5 There exist two copies of miR165 genes and six of miR166 genes in the Arabidopsis genome.5 Both miR165 and miR166 are predicted to target the transcripts of HD-ZIP III genes, which is supported by biochemical analysis6,7,8 and genetic analysis of dominant mutations of HD-ZIP III genes.7,9,10–15 An intriguing question is whether miR165 and miR166 act redundantly or play differential roles in the regulation of the HD-ZIP III genes.
Figure 1.
Alignment of mature miR165 and miR166 sequences and their complementary target sequences in the Arabidopsis HD-ZIP III mRNAs.
miR165 Overexpression Recapitulates Loss-of-Function Phenotypes of HD-ZIP III Mutants
The existence of multiple copies of miR165/166 genes in Arabidopsis provides possible multiple mechanisms in their regulation of the HD-ZIP III genes. For example, the miR165/166 genes may have different tissue expression patterns so that they regulate the functions of HD-ZIP III genes in a tissue specific manner. Although the transcripts of miR165 and miR166 have been detected in the shoot apical meristem (SAM), leaf primordia and vascular tissues in Arabidopsis,16,17,18 the expression pattern of individual miR165/166 genes is not clear. Recently, Jung and Park8 used the miR165/166 promoters fused with the glucuronidase (GUS) reporter gene to show that several miR165/166 genes exhibited different tissue-level expression patterns. It should be noted that the expression of the promoter-GUS reporter gene may not represent the endogenous gene expression pattern because the 5′ upstream sequence of a gene may not contain all the regulatory elements essential for the endogenous gene expression. Another possible mechanism of regulation by miR165/166 is that miR165 and miR166 may exhibit differential efficiency in the cleavage of HD-ZIP III transcripts due to their one-nucleotide difference (Fig. 1). One approach to test this hypothesis is to examine whether overexpression of miR165/166 could lead to downregulation of the HD-ZIP III transcripts and concomitantly recapitulate the loss-of-function phenotypes of HD-ZIP III mutants.
Overexpression by activation tagging of two Arabidopsis miR166 genes, miR166a and miR166g, has recently been shown to cause a significant reduction in the mRNA levels of three HD-ZIP III genes, ATHB-9/PHV, ATHB-14/PHB and ATHB-15.7,18 The miR166 overexpresssors exhibit an enlargement of SAM and an enhancement of vascular development, a phenotype seen in the simultaneous loss-of-function mutant of ATHB-9/PHV, ATHB-14/PHB and ATHB-15.19 The same effects were also observed by overexpression of miR166 under the cauliflower mosaic virus 35S promoter.8 Thus it appears that overexpression of miR166 only downregulates the expression of a subset of HD-ZIP III genes and recapitulates the phenotypes exhibited by simultaneous loss-of-function mutations of ATHB-9/PHV, ATHB-14/PHB and ATHB-15.
We have recently shown that overexpression of miR165 causes a reduction in the transcript levels of all five HD-ZIP III genes in Arabidopsis.20 As a result, the miR165 overexpressors exhibit a loss of SAM, an alteration in organ polarity, an inhibition in vascular development and an aberrant differentiation of interfascicular fibers. These phenotypes resemble those exhibited by the loss-of-function mutation of IFL1/REV21,22,23 and simultaneous loss-of-function mutations of IFL1/REV, ATHB-9/PHV, ATHB-14/PHB.19 These results demonstrate that overexpression of miR165 and miR166 has differential effects on the downregulation of HD-ZIP III genes and concomitantly recapitulates different phenotypes exhibited by loss-of-function mutations of HD-ZIP III genes. They suggest that the regulation of HD-ZIP III genes by miR165 and miR166 is mediated not only by their tissue specific expression but also by their differential effects on the cleavage of the target transcripts.
Overexpression of the Antisense Form of the miR165a Gene Leads to Formation of Amphivasal Vascular Bundles
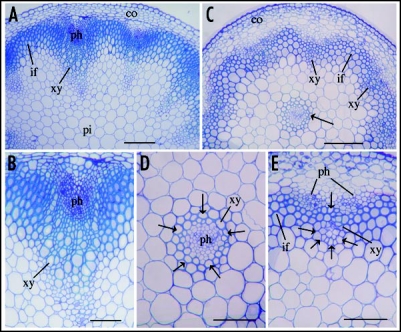
It has been shown that a point mutation in the miR165 target sequence in IFL1/REV causes an inhibition in the transcript cleavage and a high level accumulation of its full-length transcript,13 which leads to dominant phenotypes, including formation of amphivasal vascular bundles and altered organ polarity.10,13,24 To further investigate the roles of miR165 in the regulation of HD-ZIP III genes, we generated transgenic Arabidopsis plants with overexpression of the antisense form of the miR165a gene. We reasoned that antisense inhibition of miR165 should cause similar effects as the HD-ZIP III dominant mutations that occurred in the miR165 target sequence, and thus recapitulate the phenotypes exhibited by the dominant mutants. Examination of transgenic plants with antisense inhibition of miR165 revealed ectopic formation of amphivasal vascular bundles in the inflorescence stems (Fig. 2). In addition, the xylem cells in the normally placed vascular bundles also appear to be arranged in a circular pattern (Fig. 2). These results demonstrate that antisense inhibition of miR165 recapitulates the vascular phenotype exhibited by the dominant mutation of IFL1/REV and that miR165 is important in regulation of vascular patterning.
Figure 2.
Overexpression of the antisense form of the miR165a gene causes an alteration in vascular patterning. The antisense form of the miR165a gene was overexpressed under the cauliflower mosaic virus 35S promoter in wild-type Arabidopsis. Transgenic plants were selected and their inflorescence stems were sectioned and stained with toluidine blue for visualization of the vascular anatomy. (A) Section of a wild-type stem showing the regular arrangement of vascular bundles and interfascicular fibers in the stele. (B) High magnification of a wild-type vascular bundle showing the collateral vascular pattern with the xylem and phloem in parallel. (C) Section of an antisense-miR165a stem showing the aberrant formation of a vascular bundle in the middle of the stele (arrow) in addition to the normal vasculature. (D) High magnification of the misplaced vascular bundle in the antisense-miR165a stem showing an amphivasal vascular pattern with xylem (arrows) surrounding phloem. (E) High magnification of a normally placed vascular bundle in the antisense-miR165a stem showing xylem cells (arrows) arranged in a circular pattern. co, cortex; if, interfascicular fiber; ph, phloem; pi, pith; xy, xylem. Bars in (A) and (C), 197 µm and bars in (B), (D) and (E), 82 µm.
To understand the molecular mechanisms by which miR165/166 and HD-ZIP III regulate plant development, the next critical step will be to identify and study the functions of their downstream target genes. A recent global gene expression analysis revealed a link between miR165 overexpression and an alteration in the expression of genes involved in auxin signaling,20 which is consistent with the early observation that the ifl1 mutation alters polar auxin transport.23 Investigation of the inter-relationship between HD-ZIP III and their downstream genes will likely provide valuable insights in the elucidation of the complex pathways controlling vascular pattern formation, organ polarity and interfascicular fiber development in Arabidopsis.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4119
References
- 1.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7:512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Jover-Gil S, Candela H, Ponce MR. Plant microRNAs and development. Int J Dev Biol. 2005;49:733–744. doi: 10.1387/ijdb.052015sj. [DOI] [PubMed] [Google Scholar]
- 3.Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: A small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Gene Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Gene Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Kim VN, Chua NH, Park CM. MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung JH, Park CM. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta. 2007 doi: 10.1007/s00425-006-0439-1. (in press) [DOI] [PubMed] [Google Scholar]
- 9.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 10.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 11.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 12.McHale NA, Koning RE. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell. 2004;16:1730–1740. doi: 10.1105/tpc.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]
- 14.Ohashi-Ito K, Kubo M, Demura T, Fukuda H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Phsyiol. 2005;46:1646–1656. doi: 10.1093/pcp/pci180. [DOI] [PubMed] [Google Scholar]
- 15.Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, Martinez-Laborda A, Micol JL. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol. 2006;141:607–619. doi: 10.1104/pp.106.077149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- 19.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- 21.Zhong R, Taylor JJ, Ye ZH. Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell. 1997;9:2159–2170. doi: 10.1105/tpc.9.12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong R, Ye ZH. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell. 1999;11:2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong R, Ye ZH. Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 2001;126:549–563. doi: 10.1104/pp.126.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong R, Taylor JJ, Ye ZH. Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol. 1999;120:53–64. doi: 10.1104/pp.120.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]