Abstract
Calmodulin (CaM) is a versatile Ca2+-binding protein that regulates the activity of numerous effector proteins in response to Ca2+ signals. Several CaM-dependent regulatory mechanisms have been identified, including autoinhibitory domain displacement, sequestration of a ligand-binding site, active site reorganization, and target protein dimerization. We recently showed that the N- and C-lobes of animal and plant CaM isoforms could independently and sequentially bind to target peptides derived from the CaM-binding domain of Nicotiana tabacum mitogen-activated protein kinase phosphatase (NtMKP1), to form a 2:1 peptide:CaM complex. This suggests that CaM might facilitate the dimerization of NtMKP1, although the dimerization mechanism is distinct from the previously described simultaneous binding of other target peptides to CaM. The independent and sequential binding of the NtMKP1 peptides to CaM also suggests an alternative plausible scenario in which the C-lobe of CaM remains tethered to NtMKP1, and the N-lobe is free to recruit a second target protein to the complex, such as an NtMKP1 target. Thus, we hypothesize that CaM may be capable of functioning as a Ca2+-dependent adaptor or recruiter protein.
Key Words: calmodulin, calcium, EF-hand, adaptor protein, mitogen-activated protein kinase phosphatase
Calcium (Ca2+) is a dynamic secondary messenger that regulates many signaling events in both plant and animal cells. Intracellular Ca2+ transients and oscillations (Ca2+ signals) are decoded by a large superfamily of calcium-binding proteins, the most important of which is calmodulin (CaM).1–3 The prototypical CaM protein consists of four tandem helix-loop-helix “EF-hand” Ca2+-binding motifs that are divided into distinct N- and C-terminal globular lobes connected by a flexible linker. CaM proteins from all species including the single mammalian CaM and the many different plant CaM isoforms each undergo similar Ca2+-induced conformational changes involving a rearrangement of the position of its α-helices that opens distinct hydrophobic target protein-binding patches on the surface of each lobe; known as the “open” conformation (Fig. 1B). These hydrophobic patches can interact with numerous different target proteins including protein kinases, protein phosphatases, cytoskeletal proteins and other cell signaling enzymes, to regulate their activity. The closed or semi-open conformations adopted by the N- and C-lobes of Ca2+-free CaM (apo-CaM) (Fig. 1A) can also interact with another subset of proteins, to target CaM to certain cellular locations or facilitate Ca2+-independent regulatory events.1–3
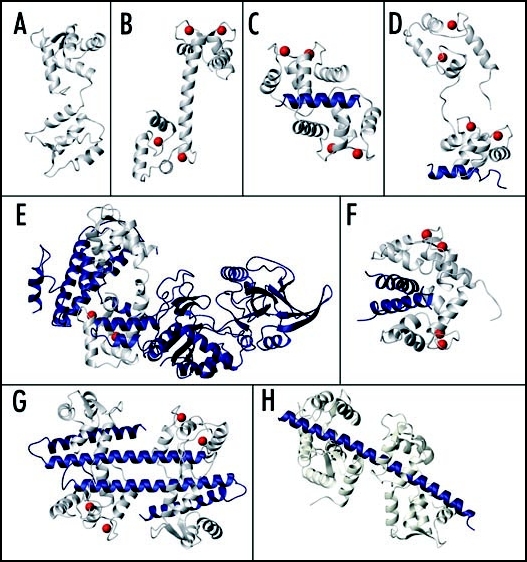
Figure 1.
Structures of CaM and CaM-target complexes. (A) apo-CaM (PDB:1DMo), (B) Ca2+-CaM (PDB:1CLL). Complexes of CaM bound to (C) CaMBD of smooth muscle myosin light chain kinase (PDB:1CDL), (D) partial CaMBD of plasma membrane Ca2+-pump C20W (PDB:1CFF), (E) the adenylyl cyclase protein from Bacillus anthracis (PDB:1K93), (F) 2 glutamate decarboxylase CaMBD's (PDB:1NWD), (G) 2 CaM proteins bound to 2 small conductance Ca2+-activated potassium channel (SK channel) CaMBD's (PDB:1G4Y), (H) 2 apo-CaM proteins bound to 2 tandem IQ motifs from murine myosin V (PDB:2IX7). In each panel CaM is shown in ivory, the target molecule is shown in blue and the Ca2+ ions bound to the N- and/or C-lobes of CaM are represented by red spheres.
The CaM-dependent regulation of target proteins can occur through numerous different mechanisms. For example, Ca2+-CaM can relieve autoinhibition by binding to a short (20–25 residue) calmodulin-binding domain (CaMBD) sequence that is adjacent to or within an autoinhibitory region of the enzyme (Fig. 2A).3 Numerous structures of these Ca2+-CaM-CaMBD complexes have been reported, which reveal a characteristic “wrap-around” binding mode (Fig. 1C). Typically the CaM C-lobe binds with high affinity to a Trp residue within the N-terminal part of the target sequence, and the flexible central linker allows the N-lobe to pivot and bind to a second bulky hydrophobic “anchor” residue within the C-terminal part of the target sequence.3 Truncation of this second anchor residue can lead to binding of only one CaM domain and an extended CaM conformation (Fig. 1D).4,5 Studies with plant CaM isoforms having mutations to non-CaMBD-coordinating residues have also suggested that a secondary binding interface exists on the opposite surface of the CaM protein which also contributes to the activation of some of these target enzymes.6,7
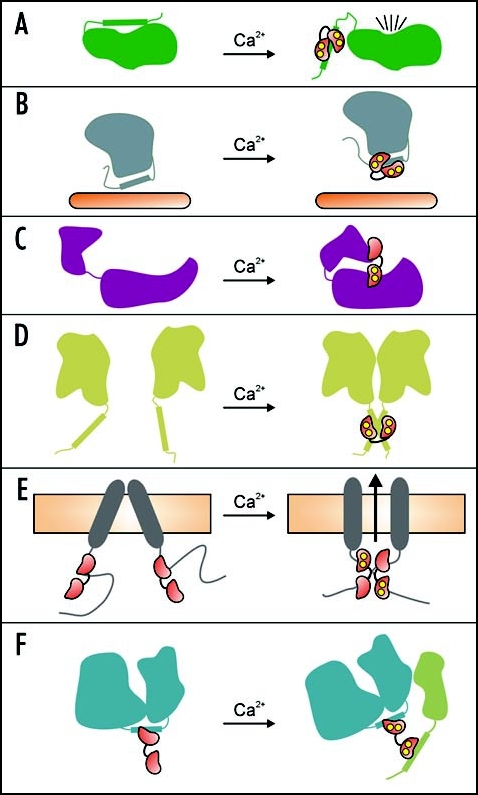
Figure 2.
Schematic model for the various mechanisms of CaM-dependent target regulation. (A) autoinhibitory domain displacement, (B) sequestering of a ligand binding site, (C) active-site reorganization, (D) CaM-induced target protein dimerization (1:2 complex), (E) CaM-induced target protein dimerization (2:2 complex), (F) hypothesized model for CaM acting as an adaptor/recruiter protein. In each panel CaM is shown as a red dumbbell shaped molecule with Ca2+ ions represented by yellow circles, and the target proteins are shown in various colors. See the text for details on each model.
Another regulatory mechanism involving Ca2+-CaM-binding to a single contiguous CaMBD sequence may occur with the potato kinesin-like CaM-binding protein (KCBP)8 as well as some plant cyclic-nucleotide gated channels (CNGC's).9 In both cases the Ca2+-CaM binding site on the target protein overlaps with the respective ligand binding site, and thus the binding of KCBP to microtubules or the binding of cyclic nucleotide monophosphates to CNGC's may be prevented by interaction with Ca2+-CaM (Fig. 2B). In a variation on this mechanism, CaM can bind to the cytoplasmic juxtamembrane region of the human epidermal growth factor receptor and sequester a threonine residue which is a specific phosphorylation target of protein kinase C (PKC). CaM-binding inhibits PKC phosphorylation of this threonine, and PKC phosphorylation inhibits CaM-binding.10
There are also several examples of CaM-target interactions where the N- and C-lobes bind to noncontiguous target protein regions, and play distinct roles in target regulation. The structures of a CaM-activated adenylyl cyclase from Bacillus anthracis with and without bound CaM shows how the N- and C-lobes of CaM can bind two distant regions of the adenylyl cyclase enzyme and induce a conformation reorganization that creates the enzyme's active site (Figs. 1E and 2C).11 An interesting feature of this interaction is that the CaM N-lobe remains Ca2+-free and in a closed conformation, while the C-lobe is in a canonical Ca2+-bound open conformation. Indeed, Ca2+-binding to the C-lobe but not N-lobe is required for activation of the adenylyl cyclase.12
The N- and C-lobes of Ca2+-CaM can also each simultaneously bind to identical peptides derived from the petunia glutamate decarboxylase (GAD) enzyme to form a 1:2 Ca2+-CaM:GAD complex (Fig. 1F).13,14 This suggests that Ca2+-CaM-induced target protein dimerization may be another way in which CaM can regulate target proteins (Fig. 2D). CaM-dependent dimerization has also been shown to regulate the activity of small conductance Ca2+-activated K+ channels (SK channel), although in this case a novel 2:2 CaM:SK channel complex is formed (Figs. 1G and 2E).15 This structure is also unique because Ca2+ is bound to the “lower affinity” N-lobe EF-hands, but not to the “higher affinity” C-lobe EF-hands of CaM.
In addition to the SK channel, CaM can regulate voltage-gated sodium channels, voltage-gated calcium channels, as well as ryanodine-sensitive calcium release channels.16 With these channels CaM typically binds in complex Ca2+-dependent and Ca2+-independent ways to several noncontiguous target sequences in the same protein, and often to so-called IQ motifs (IQXXXRGXXXR). IQ motifs are generally thought to be constitutive apo-CaM binding sites which retain CaM under resting (low [Ca2+]) cellular conditions to ensure a rapid response to Ca2+-stimuli.17 However many IQ motifs can also bind specifically to Ca2+-CaM or to both apo-CaM and Ca2+-CaM. Structures of some Ca2+-CaM-IQ domain complexes have revealed wrap-around binding modes, albeit with differences in lobe and peptide orientation compared to other complexes.18–20 For a discussion about the mechanisms of CaM-dependent ion channel regulation (see ref. 16). A very recent crystal structure of apo-CaM bound to an IQ domain from myosin V (Fig. 1H) has also revealed yet another variation on the wrap-around binding mode, where the apo-C-lobe of CaM adopts a semi-open conformation and forms numerous interactions with the target sequence, while the apo-N-lobe adopts a closed conformation and forms weaker interactions with the IQ domain.21
Using several biophysical techniques we recently characterized the interaction between CaM isoforms (mammalian CaM, soybean CaM isoforms SCaM-1 and SCaM-4) and a novel CaMBD derived from the Nicotiana tabacum mitogen-activated protein kinase phosphatase (NtMKP1).22 The NtMKP1 protein was initially identified as a CaM-binding protein by Ohashi and coworkers,23 and the same group recently showed that CaM-binding NtMKP1 homologs are also present in other plant species as well.24 We found that each CaM isoform was capable of binding to the NtMKP1 CaMBD in the absence of Ca2+ using only the apo-C-lobe, with the primary binding site consisting of NtMKP1 residues N438 - S449, and additional C-terminal residues G450 - K460 enhancing the overall binding affinity (Kd ∼10−5 M). In the presence of Ca2+, a 1:1 complex could be formed with the CaM C-lobe having significantly increased affinity for the N438 - S449 region of NtMKP1 (Kd 10−7 − 10−10 M). However, the Ca2+-loaded CaM N-lobe interacted only very weakly with the C-terminal NtMKP1 sequence in this 1:1 complex, despite an abundance of seemingly suitable hydrophobic “anchor” residues in this region. Interestingly, the addition of more peptide triggered the independent binding of a second NtMKP1 peptide to the Ca2+-CaM N-lobe (Kd 10−5 − 10−6 M) to form a 1:2 Ca2+-CaM:NtMKP1 complex. As with GAD, these results suggest that CaM is capable of facilitating the dimerization of NtMKP1, although the independent and sequential NtMKP1 peptide binding to the C- and N-lobes markedly distinguishes the CaM-NtMKP1 interaction from the simultaneous high-affinity binding of 2 GAD CaMBD's to CaM.
While our NtMKP1 study was ongoing, Ohashi and coworkers reported that CaM is incapable of stimulating the phosphatase activity of the NtMKP1 enzyme, thereby implying that the CaM-NtMKP1 interaction is necessary for something other than direct enzyme regulation.25 The independent and sequential binding of the NtMKP1 fragments to the Ca2+-saturated C- and then N-lobes of CaM observed in our study suggests a plausible situation in which the C-lobe of CaM is tightly bound to NtMKP1, leaving the N-lobe free to recruit a different target protein to the complex, for example, a NtMKP1 protein substrate. Therefore, CaM may be capable of acting as an adaptor or recruiter protein, which would add yet another mechanism of target regulation to CaM's repertoire (Fig. 2F). In addition to NtMKP1 peptides, the isolated N-lobe of CaM is capable of binding to other CaMBD peptides26,27 as well as intact target proteins,28 increasing the likelihood that the N-lobe could serve as a recruiter domain. The pre-association of the apo-C-lobe of CaM with NtMKP1 under resting conditions would also ensure a rapid response response to Ca2+-stimuli, since CaM would only need to recruit one rather than both protein targets.
Although the ability of CaM to act as an adaptor protein in vivo has not yet been demonstrated, there are examples of related EF-hand proteins acting as adaptor proteins, including centrin29 and calcium- and integrin-binding protein 1.30 With the abundance of poorly characterized CaM-binding proteins in plants, many of which have CaMBD's with little sequence resemblance to the better characterized motifs in animals1 it seems likely that sequences will be identified which bind preferentially to the CaM N-lobe. Considering the incredible assortment of known CaM interaction modes and regulatory mechanisms, many of which have only been identified within the last decade, it is likely only a matter of time before CaM is proven to function as an adaptor protein in vivo.
Abbreviations
- Cam
calmodulin
- CaMBD
calmodulin-binding domain
- KCBP
kinesin-like CaM-binding protein
- CNGC's
cyclic-nucleotide gated channels
- PKC
protein kinase C
- GAD
glutamate decarboxylase
- SK channel
small conductance Ca2+-activated K+ channels
- NtMKP1
Nicotiana tabacum mitogen-activated protein kinase phosphatase NtMKP1
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4155
References
- 1.Bouche N, Yellin A, Snedden WA, Fromm H. Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol. 2005;56:435–466. doi: 10.1146/annurev.arplant.56.032604.144224. [DOI] [PubMed] [Google Scholar]
- 2.Jurado LA, Chockalingam PS, Jarrett HW. Apocalmodulin. Physiol Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- 3.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 4.Elshorst B, Hennig M, Forsterling H, Diener A, Maurer M, Schulte P, Schwalbe H, Griesinger C, Krebs J, Schmid H, Vorherr T, Carafoli E. NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry. 1999;38:12320–12332. doi: 10.1021/bi9908235. [DOI] [PubMed] [Google Scholar]
- 5.Kataoka M, Head JF, Vorherr T, Krebs J, Carafoli E. Small-angle X-ray scattering study of calmodulin bound to two peptides corresponding to parts of the calmodulin-binding domain of the plasma membrane Ca2+ pump. Biochemistry. 1991;30:6247–6251. doi: 10.1021/bi00239a024. [DOI] [PubMed] [Google Scholar]
- 6.Lee SH, Seo HY, Kim JC, Heo WD, Chung WS, Lee KJ, Kim MC, Cheong YH, Choi JY, Lim CO, Cho MJ. Differential activation of NAD kinase by plant calmodulin isoforms: The critical role of domain I. J Biol Chem. 1997;272:9252–9259. doi: 10.1074/jbc.272.14.9252. [DOI] [PubMed] [Google Scholar]
- 7.Van Lierop JE, Wilson DP, Davis JP, Tikunova S, Sutherland C, Walsh MP, Johnson JD. Activation of smooth muscle myosin light chain kinase by calmodulin: Role of LYS(30) and GLY(40) J Biol Chem. 2002;277:6550–6558. doi: 10.1074/jbc.M111404200. [DOI] [PubMed] [Google Scholar]
- 8.Vinogradova MV, Reddy VS, Reddy AS, Sablin EP, Fletterick RJ. Crystal structure of kinesin regulated by Ca(2+)-calmodulin. J Biol Chem. 2004;279:23504–23509. doi: 10.1074/jbc.M400741200. [DOI] [PubMed] [Google Scholar]
- 9.Arazi T, Kaplan B, Fromm H. A high-affinity calmodulin-binding site in a tobacco plasma-membrane channel protein coincides with a characteristic element of cyclic nucleotide-binding domains. Plant Mol Biol. 2000;42:591–601. doi: 10.1023/a:1006345302589. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Nieto J, Villalobo A. The human epidermal growth factor receptor contains a juxtamembrane calmodulin-binding site. Biochemistry. 1998;37:227–236. doi: 10.1021/bi971765v. [DOI] [PubMed] [Google Scholar]
- 11.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang WJ. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 12.Drum CL, Yan SZ, Sarac R, Mabuchi Y, Beckingham K, Bohm A, Grabarek Z, Tang WJ. An extended conformation of calmodulin induces interactions between the structural domains of adenylyl cyclase from Bacillus anthracis to promote catalysis. J Biol Chem. 2000;275:36334–36340. doi: 10.1074/jbc.M004778200. [DOI] [PubMed] [Google Scholar]
- 13.Yuan T, Vogel HJ. Calcium-calmodulin-induced dimerization of the carboxyl-terminal domain from petunia glutamate decarboxylase: A novel calmodulin-peptide interaction motif. J Biol Chem. 1998;273:30328–30335. doi: 10.1074/jbc.273.46.30328. [DOI] [PubMed] [Google Scholar]
- 14.Yap KL, Yuan T, Mal TK, Vogel HJ, Ikura M. Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J Mol Biol. 2003;328:193–204. doi: 10.1016/s0022-2836(03)00271-7. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 16.Shah VN, Chagot B, Chazin WJ. Calcium-dependent regulation of ion channels. Calcium Bind Prot. 2007;1:203–212. [PMC free article] [PubMed] [Google Scholar]
- 17.Bahler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 18.Fallon JL, Halling DB, Hamilton SL, Quiocho FA. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure. 2005;13:1881–1886. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Maximciuc AA, Putkey JA, Shamoo Y, Mackenzie KR. Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure. 2006;14:1547–1556. doi: 10.1016/j.str.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Van Petegem F, Chatelain FC, Minor DL., Jr Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat Struct Mol Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houdusse A, Gaucher JF, Krementsova E, Mui S, Trybus KM, Cohen C. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc Natl Acad Sci USA. 2006;103:19326–19331. doi: 10.1073/pnas.0609436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainaldi M, Yamniuk AP, Murase T, Vogel HJ. Calcium-dependent and -independent binding of soybean calmodulin isoforms to the calmodulin binding domain of tobacco MAPK phosphatase-1. J Biol Chem. 2007;282:6031–6042. doi: 10.1074/jbc.M608970200. [DOI] [PubMed] [Google Scholar]
- 23.Yamakawa H, Katou S, Seo S, Mitsuhara I, Kamada H, Ohashi Y. Plant MAPK phosphatase interacts with calmodulins. J Biol Chem. 2004;279:928–936. doi: 10.1074/jbc.M310277200. [DOI] [PubMed] [Google Scholar]
- 24.Katou S, Kuroda K, Seo S, Yanagawa Y, Tsuge T, Yamazaki M, Miyao A, Hirochika H, Ohashi Y. A calmodulin-binding mitogen-activated protein kinase phosphatase is induced by wounding and regulates the activities of stress-related mitogen-activated protein kinases in rice. Plant Cell Physiol. 2007;48:332–344. doi: 10.1093/pcp/pcm007. [DOI] [PubMed] [Google Scholar]
- 25.Katou S, Karita E, Yamakawa H, Seo S, Mitsuhara I, Kuchitsu K, Ohashi Y. Catalytic activation of the plant MAPK phosphatase NtMKP1 by its physiological substrate salicylic acid-induced protein kinase but not by calmodulins. J Biol Chem. 2005;280:39569–39581. doi: 10.1074/jbc.M508115200. [DOI] [PubMed] [Google Scholar]
- 26.Shuman CF, Jiji R, Kerfeldt KS, Linse S. Reconstitution of calmodulin from domains and subdomains: Influence of target peptide. J Mol Biol. 2006;358:870–881. doi: 10.1016/j.jmb.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Yuan T, Tencza S, Mietzner TA, Montelaro RC, Vogel HJ. Calmodulin binding properties of peptide analogues and fragments of the calmodulin-binding domain of simian immunodeficiency virus transmembrane glycoprotein 41. Biopolymers. 2001;58:50–62. doi: 10.1002/1097-0282(200101)58:1<50::AID-BIP60>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 28.Medvedeva MV, Kolobova EA, Wang P, Gusev NB. Interaction of proteolytic fragments of calmodulin with caldesmon and calponin. Biochem J. 1996;315:1021–1026. doi: 10.1042/bj3151021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan JH, Bunick CG, Hu H, Fagan PA, Meyn SM, Chazin WJ. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J Biol Chem. 2006;281:2876–2881. doi: 10.1074/jbc.M509886200. [DOI] [PubMed] [Google Scholar]
- 30.Tsuboi S, Nonoyama S, Ochs HD. Wiskott-Aldrich syndrome protein is involved in alphaIIb beta3-mediated cell adhesion. EMBO J. 2006;7:506–511. doi: 10.1038/sj.embor.7400665. [DOI] [PMC free article] [PubMed] [Google Scholar]