Abstract
Reactive oxygen species have been suggested to play a signaling role in seed dormancy alleviation. When sunflower seeds become able to fully germinate during dry after-ripening, they accumulate high amount of hydrogen peroxide and exhibit a low detoxifying ability through catalase, resulting from the decrease in CATA1 transcript. ROS accumulation entails oxidative modification of soluble and storage proteins through carbonylation, which suggests that this process might play an important role in plant developmental processes. However other oxidative signaling pathways cannot be excluded. For example, a cDNA-AFLP study shows that seed after-ripening is also associated with changes in gene expression and that changes in ROS content during seed imbibition are also related to changes in expression pattern.
Key Words: seed dormancy, reactive oxygen species, after-ripening, carbonylation, transcription
Introduction
Seed dormancy release is a very fascinating process that consists in a dramatic change in seed physiology, since seeds progressively acquire the ability to germinate in conditions which prevent germination in the dormant state. In addition this phenomenon can occur for many species during dry storage (after-ripening), although there is no free water available in the tissue for theoretically sustaining any metabolic activity. Despite a huge number of studies on this topic, the cellular and molecular basis of this mechanism is still poorly understood. We recently demonstrated that reactive oxygen species (ROS) are likely to play a signaling function in seed dormancy alleviation, and that they target specific changes in protein oxidation, including during after-ripening. These data bring novel insights into the understanding of seed dormancy that we wish to underline in the present paper.
ROS are a Signal in Seed Dormancy Release
Our study demonstrates that ROS, i.e., superoxide anions and hydrogen peroxide, and peroxidation products, i.e., malondialdehyde, accumulated in embryonic axes of sunflower seeds during dry storage. These data suggest that ROS production is a continuous phenomenon which is initiated immediately after harvest. During seed development on the plant, ROS content regularly declines till harvest1,2 therefore being low at harvest time, and it increases again after seed dispersion from the mother plant. We propose that this increase would be beneficial for the seed in a first step, allowing elimination of dormancy. However, upon prolonged storage or storage under inappropriate conditions (temperature, relative humidity) the ROS content could reach a critical level, being harmful for the seed and inducing an oxidative stress and related damage. However, in both cases, the role of ROS, either beneficial or detrimental, is unraveled when seeds get imbibed. Interestingly, the amount of CATA1 transcript, one of the four genes coding for catalase in sunflower,1 declines during dry after-ripening, raising the hypothesis that the H2O2 scavenging capacity of nondormant seeds is low during the early phase of imbibition. In perfect agreement with this, nondormant imbibed seeds exhibit much higher H2O2 amounts than the dormant ones (Table 1). However, as germination progresses, there is a reinduction of the CATA1 gene occurring after 16 h of imbibition. The importance of ROS as a signal for germination is emphasized by the use of methylviologen (MV), a ROS generating compound breaking seed dormancy in sunflower. The main perspective of this finding will be also to draw a comprehensive and integrative model of dormancy alleviation in the imbibed state with regards to the interplay between ROS and plant hormones, which has been demonstrated in several processes. For example, Desikan et al.3 showed that the ethylene receptor ETR1 was mediating H2O2 signal in stomatal guard cells, and interestingly, ethylene is also known to alleviate sunflower seed dormancy.4
Table 1.
Hydrogen peroxide content and CATA1 gene expression in axes isolated from dormant and non-dormant sunflower seeds before imbibition (dry) or after 8 and 16 h of imbibition on water at 10°C.
| Duration of imbibition (h) | H2O2 content (µmol g DW-1) | CATA1 expression (relative units) | ||
| dormant | non-dormant | dormant | non-dormant | |
| 0 (dry) | 1.37 ± 0.21 | 2.10 ± 0.14 | 100 | 46.0 |
| 8 | 1.43 ± 0.35 | 4.52 ± 0.65 | 48.0 | 44.6 |
| 16 | 1.50 ± 0.22 | 8.22 ± 1.26 | 59.3 | 114 |
Hydrogen peroxide content and CATA1 gene expression in axes isolated from dormant and non-dormant sunflower seeds before imbibition (dry) or after 8 and 16 h of imbibition on water at 10°C.. Values are means of five replicates ± SD (H2O2). CATA1 expression was determined by northern blot and is % of the expression determined in dry dormant seeds (means of three measurements).
What is the Relevance of Protein Carbonylation in Seeds?
Protein carbonylation is an irreversible reaction which inhibits the activities and/or increases the vulnerability to proteases of the proteins targeted to oxidation damage.5,6 In humans, it plays a significant role in the progression of several diseases such as Alzheimer's or diabetes,7 and carbonylated proteins are considered as a potent biomarker of oxidative stress, as well as in plants.8 In Arabidopsis, Johansson et al.9 showed that carbonylation could be developmentally regulated and Job et al.10 also demonstrated that this process occurred during the germination of fully viable seeds. Interestingly our work demonstrated that specific carbonylation was involved in seed dormancy alleviation, either in the dry state or during imbibition, although the nature of the oxidized proteins differed in the two cases. The specificity of carbonylation towards soluble proteins was shown in Oracz et al.11 but is further emphasized by (Fig. 1), which displays 1D gels of storage proteins and the DNP-assay for detection of protein carbonyls. The gels displayed major protein bands of about 51, 35, 29 and 22 kDa which were assigned by LC/MS-MS analyses to acidic and basic chains of 11S globulins (being called helianthinins in sunflower). Four distinct bands of ca 16, 35, 50 and 60 kDa carbonylated proteins were detected but their intensity was very similar in both dormant and nondormant axes (Fig. 1), which suggests that seed imbibition increased the susceptibility of storage proteins to proteolysis, independently of the subsequent completion of germination. This would allow the imbibed nongerminating dormant seeds to sustain a minimal metabolic activity during imbibition, a finding that is in accordance with the recent results of Chibani et al.12 Therefore, in addition to the data provided by Job et al.,10 our results demonstrate that carbonylation can play a regulatory role and should not always be regarded as an unfavorable mechanism in developmental processes.
Figure 1.
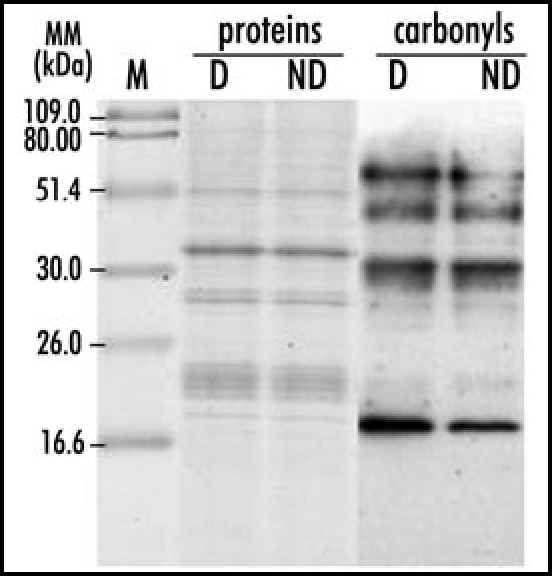
1D PAGE of oxidatively modified storage proteins from axes of dry dormant and nondormant sunflower seeds. Protein stain (proteins) and anti-DNP (2,4-dinitrophenylhydrazone) immunoassay (carbonyls) are shown. D, dormant; ND, nondormant; M, markers; MM, molecular mass.
Is the Proteome the only Target of ROS in Seed Dormancy Alleviation?
Further studies have stated that alleviation of seed dormancy, including in the dry state, was mainly related to a change in the seed transcriptome.13–15 Although we propose ROS production and proteome oxidation as a key regulatory mechanism in the breaking of dormancy, we consider that the regulation of this phenomenon is multifactorial and requires a coordinated activation of various molecular mechanisms. Taking the effect of H2O2 on gene expression into consideration,16 we investigated the putative involvement of transcriptome in sunflower seed dormancy release in the dry state, using methylviologen (which generates ROS) and cyanide, which allows the germination of dormant seed by triggering similar protein carbonylation as methylviologen.11 Using cDNA-AFLP, we observed that, with a combination of 15 primers, 16 identical reproducible transcription derived fragments (TDF) were found in axes of dormant sunflower seeds treated by either cyanide or MV, whereas five and ten combinations, respectively, were fully specific of each compound (Table 2). These results demonstrate that alleviation of sunflower seed dormancy is also related to changes in gene expression, which may be caused by ROS accumulation. As for proteome oxidation, it is worth to note that the effects of MV and cyanide on gene expression share some homology, highlighting again that their mechanisms of action are probably very close, involving ROS signaling. We also observed that 11 out of 78 TDF were found in nondormant seeds after after-ripening (data not shown) showing that changes in gene expression may occur in the dry state, as already suggested by Leubner-Metzer,17 Bove et al.13 and Leymarie et al.15
Table 2.
cDNA-AFLP expression pattern of transcript-derived fragments (TDF) identified as polymorphic in embryonic axes of dormant (D) and non-dormant (ND) seeds after 3h of imbibition on water or in the presence of hydrogen cyanide 1 mM (CN) or methylviologen 1 mM (MV) at 10°C.
| cDNA-AFLP expression in | Amount of TDF | Specificity | |||
| D + water | ND + water | D + CN | D + MV | ||
| + | − | − | − | 2 | D |
| − | + | − | − | 9 | ND |
| − | − | + | + | 15 | CN and MV |
| + | + | − | − | 1 | CN and MV |
| − | − | + | − | 5 | CN |
| − | − | − | + | 10 | MV |
| − | + | − | + | 2 | ND and MV |
| − | + | + | − | 2 | ND and CN |
| − | + | + | + | 1 | germination |
| + | + | + | + | 4 | none |
The specificity of the expression pattern is indicated in the right column. The number of TDF observed among the 51 primer combinations tested is indicated.
Perspective
The discovery that ROS play a signaling role in seed dormancy release opens new perspectives in this field of plant science. We think that further work should pay more attention to the mechanisms of ROS production in the dry state, which would help to better appreciate the possible basis of seed life in anhydrobiosis, including during seed aging. In imbibed seeds, the interplay of hormones, and particularly ABA, with ROS needs to be addressed in order to get a more comprehensive view of the cellular basis of seed dormancy. At last, other mechanisms of ROS-mediated signaling pathways should be also investigated; they may include other oxidative modification of proteins, changes in the redox status, lipid and sugar signaling or alteration of DNA structure.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4460
References
- 1.Bailly C, Leymarie J, Lehner A, Rousseau S, Come D, Corbineau F. Catalase activity and expression in developing sunflower seeds as related to drying. J Exp Bot. 2004;55:475–483. doi: 10.1093/jxb/erh050. [DOI] [PubMed] [Google Scholar]
- 2.Bailly C. Active oxygen species and antioxidants in seed biology. Seed Sci Res. 2004;14:93–107. [Google Scholar]
- 3.Desikan R, Hancock JT, Bright J, Harrison J, Weir I, Hooley R, Neill SJ. A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 2005;137:831–834. doi: 10.1104/pp.104.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbineau F, Bagniol S, Côme D. Sunflower (Helianthus annuus L.) seed dormancy and its regulation by ethylene. Isr J Bot. 1990;39:313–325. [Google Scholar]
- 5.Berlett BS, Stadtman ER. Protein oxidation in aging, disease and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 6.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen AT, Donaldson RP. Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch Biochem Biophy. 2005;439:25–31. doi: 10.1016/j.abb.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Johansson E, Olsson O, Nyström T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem. 2004;279:22204–22208. doi: 10.1074/jbc.M402652200. [DOI] [PubMed] [Google Scholar]
- 10.Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005;138:790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oracz K, Bouteau HE, Farrant JM, Cooper K, Belghazi M, Job C, Job D, Corbineau F, Bailly C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007;50:452–465. doi: 10.1111/j.1365-313X.2007.03063.x. [DOI] [PubMed] [Google Scholar]
- 12.Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 2007;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bove J, Lucas P, Godin B, Oge L, Jullien M, Grappin P. Gene expression analysis by cDNA-AFLP highlights a set of new signaling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia. Plant Mol Biol. 2005;57:593–612. doi: 10.1007/s11103-005-0953-8. [DOI] [PubMed] [Google Scholar]
- 14.Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 15.Leymarie J, Bruneaux E, Gibot-Leclerc S, Corbineau F. Identification of transcripts potentially involved in barley seed germination and dormancy using cDNA-AFLP. J Exp Bot. 2007;58:425–437. doi: 10.1093/jxb/erl211. [DOI] [PubMed] [Google Scholar]
- 16.Desikan R, AH Mackerness S, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leubner-Metzer G. Beta-1,3-Glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. Plant J. 2005;41:133–145. doi: 10.1111/j.1365-313X.2004.02284.x. [DOI] [PubMed] [Google Scholar]


