Abstract
In the auxin signal transduction, two protein families, Aux/IAAs and auxin response factors, play a crucial role just downstream of auxin F-box receptors. Distinct and overlapping phenotypes of the dominant Aux/IAA mutants suggest some functional differentiation of the Aux/IAA genes in auxin signaling. Taking advantage of unique phenotypes of the msg2/iaa19 mutants, we carried out promoter-exchange experiments, where cDNA of the msg2, axr2/iaa7 or slr/iaa14 gene was driven by the MSG2 or AXR2 promoter. The cDNAs were translationally fused to the green fluorescent protein gene to measure levels of expressed protein. Results showed that many abnormal phenotypes of the dominant Aux/IAA mutants were governed by their promoter activity, but some were dependent on their gene products. The latter result highlights the possible importance of Aux/IAA protein level controled by auxin F-box receptors.
Key Words: auxin signal transduction, auxin response factor, Aux/IAA protein, gene expression, protein degradation
Auxin exerts many physiological responses in different tissues, which has been a puzzle since its discovery in the 1920's. The discovery of two protein families, auxin response factors (ARFs) and Aux/IAAs, in the late 1990's was, thus, epoch-making because each physiological response might result from combinatorial interaction between a subset of the ARF and Aux/IAA families, which consist of 23 and 29 proteins in Arabidopsis, respectively.1,2 Consequently, each Aux/IAA gene is thought to be differentiated in their physiological function, at least to some extent. Consistent with this idea, dominant Arabidopsis mutants of the Aux/IAA genes show both distinct and overlapping phenotypes. The next question is: What makes each Aux/IAA differentiated on a molecular level?
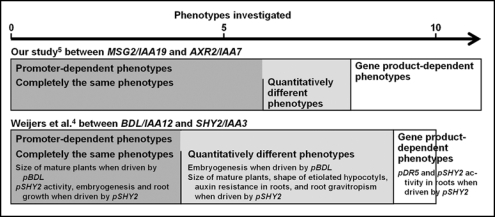
Theoretically there would be two extremes for this question. One is that the function of Aux/IAAs is solely decided by their expression pattern. In this case, all the dominantly mutated Aux/IAA proteins (mAux/IAAs) would produce similar defects if expressed in the same tissue. The other is that each Aux/IAA could interact with a distinct set of ARFs, leading to phenotypic defects characteristic to the repressed ARFs. In this case, each mAux/IAAs should induce qualitatively different defects even if expressed by the same promoter. This question has been addressed before by Knox et al.3 and Weijers et al.4 by the use of the promoter-swapping strategy. In the latest issue of Plant Physiology we also reported our results on this question5 by taking advantage of the msg2/iaa19 mutants, which exhibit fewer defects than the other dominant Aux/IAA mutants.6,7 Figure 1 summarizes our results as well as those of Weijers et al.4 In our experiments, cDNA of msg2, axr2/iss78 or slr-1/iaa149 was driven by the MSG2 or AXR2 promoter. Weijers et al. expressed the bdl/iaa1210 or shy2/iaa311 cDNA by the BDL or SHY2 promoters.
Figure 1.
Promoter- and gene product-dependent phenotypes of the dominant Aux/IAA mutants as revealed by promoter-exchange experiments. pBDL represents promoter of the BDL gene.
Of the 21 determined phenotypes in total, mAux/IAA proteins induce the same or qualitatively similar defects in 17 phenotypes (Fig. 1). In four cases, however, mAux/IAA did exert qualitatively different phenotypes, even when driven by the same promoter. This clearly shows that physiological function of mAux/IAA was determined by both the pattern of gene expression and the properties of gene products, but that gene expression may have a primary role. This conclusion is essentially the same as that reached by the previous study.4
The importance of gene expression has been widely recognized in studies of gene function. Thus, it would be surprising if each mAux/IAA protein had distinct characteristics, and the next question would be: What properties of the Aux/IAA proteins make them distinct from each other? The Aux/IAA proteins consist of three conserved regions, domain I, domain II and the carboxy-terminal domain (CTD). Domain II is a recognition site for auxin F-box receptors (AFBs).12–14 AFBs ubiquitinate Aux/IAAs after auxin perception, leading to degradation of Aux/IAA. This relieves ARFs from repression of their transcriptional activities, which ultimately results in auxin responses. Therefore, properties of Aux/IAAs are likely determined by the binding constants for ARFs through CTD and for AFBs through domain II. Because strength of interaction between Aux/IAAs and ARFs appears to be similar for pairs investigated so far with yeast two-hybrid assay4,6,9,10,15,16 or fluorescence cross-correlation spectroscopy,17 binding constants between Aux/IAAs and AFBs may be variable. In fact, when driven by the same AXR2 or MSG2 promoter, a protein level of msg2-1 estimated from fluorescence intensity of green fluorescent protein fused to msg2-1 was much lower than that of axr2-1,5 suggesting that msg2-1 has a higher affinity to AFBs. This difference may cause a few msg2-specific defects independent of the promoter activities (Table 1, underlined). Even in the case where mAux/IAAs exhibited quantitatively different phenotypes, msg2-1 exerted weaker defects than did slr-1 and axr2-1 (Table 1, shaded). This may also be due to a lower msg2 level than the other two mAux/IAAs. Quantitative determination of the interaction between Aux/IAAs and AFBs will be needed to further understand functional differentiation of the Aux/IAA family.
Table 1.
Shared and differentiated functions among the dominantly mutated Aux/IAA proteins
| Phenotype | Degree of Defects | Target Auxin Response Factors (ARF) (Putative) | ||||
| Dominant Mutants | Driven by MSG2 Promoter | Driven by AXR2 Promoter | ||||
| Size of nature plants | wt1) = msg2 < slr < axr2 | wt = | msg2 = slr = axr2* | wt = | msg2 < slr = axr2 | ARF6 + ARF818 |
| Embryogenesis | wt = msg2 = slr = axr2 +NPH4/ARF716 | wt = | msg2 < slr = axr2 | wt = | msg2 = slr = axr2* | MP/ARF5 |
| Shape of etiolated hypocotyls | wt ≈ msg2 < slr, axr22) | wt ≈ | msg2 = slr = axr2 | wt ≈ | msg2 < slr, axr22) | ARF6 + ARF818 |
| Hypocotyl gravitropism | wt < msg2 < slr < axr2 | wt < | msg2< slr = axr2 | wt < | msg2< slr = axr2 | NPH4 + ARF1919,20 |
| Lateral root formation | wt < msg2, axr22) < slr | wt < | msg2 = slr = axr2 | ND1) | NPH4 + ARF1920,21 | |
| Root gravitropism | wt = msg2 < slr = axr2 | wt = | msg2 = slr = axr2* | ND | NPH4 + ARF1920 | |
| Root hair formation | wt = msg2 < slr = axr2 | wt = | msg2 = slr = axr2* | ND | ? | |
This shows that MSG2 or AXR2 does not express in a tissue critical for the phenotype. Proteins appearing in a black background show the same function among the mutated Aux/IAA proteins; those in a grey background show quantitative differences in function among them; underlined names show qualitative differences;
wt, wild type; ND, not determined;
Two mutants exhibit qualitatively different phenotypes.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4264
References
- 1.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knox K, Grierson CS, Leyser O. AXR3 and SHY2 interact to regulate root hair development. Development. 2004;130:5769–5777. doi: 10.1242/dev.00659. [DOI] [PubMed] [Google Scholar]
- 4.Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments between MSG2/IAA19, AXR2/IAA3 and SLR/IAA14. Plant Physiol. 2007;144:187–196. doi: 10.1104/pp.107.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito K, Watahiki MK, Yamamoto KT. Differential expression of the auxin primary-response gene MASSUGU2/IAA19 during tropic responses of Arabidopsis hypocotyls. Physiol Plant. 2007;130:148–156. [Google Scholar]
- 8.Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005;44:382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- 10.Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Q, Uhlir NJ, Reed J. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 13.Kepinski S, Leyser O. The Arabidopsis TIR1 protein is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 14.Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 15.Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen T, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 17.Muto H, Nagao I, Demura T, Fukuda H, Kinjo M, Yamamoto KT. Fluorescence cross-correlation analyses of molecular interaction between Aux/IAA protein and protein-protein interaction domain of auxin response factors of Arabidopsis expressed in HeLa cells. Plant Cell Physiol. 2006;47:1095–1101. doi: 10.1093/pcp/pcj080. [DOI] [PubMed] [Google Scholar]
- 18.Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 19.Watahiki MK, Yamamoto KT. The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol. 1997;115:419–426. doi: 10.1104/pp.115.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologisa A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]