Abstract
Shoot development is regulated by specific gene expression programs depending on the interplay between transcription factors and growth hormones that function in specific domains of the meristem and lateral organs. Functional relationship between different regulators is not clearly established. In the May issue of Plant Physiology (2007) we have shown that Wuschel-like Homeobox3 and YABBY3 are coexpressed in the leaf primordia and young leaves, and that WOX3 functions as a transcriptional repressor of YAB3. Overexpression of WOX3 or downregulation of YAB3 induced ectopic expression of Knotted 1-like homeobox1 genes in leaves and consequently produced a phenotype similar to plants ectopically expressing KNOXI genes. In a parallel work published in the same issue, we have shown that another YABBY gene, YAB1, which is expressed in the same domains as YAB3 or WOX3, binds to the gibberellic acid responsive element and is involved in the feedback regulation of gibberellin biosynthesis. Our study demonstrates that rice YABBY genes are involved in two pathways to control cell division and differentiation during leaf formation and growth and establishes a rice shoot developmental regulatory hierarchy involving WOX3, YAB3, KNOX1 and gibberellin.
Key Words: rice, shoot, meristem, Wuschel-like homeobox, YABBY, KNOX1, gibberellin, feedback regulation and repression
The development of shoot apical meristem (SAM) is regulated by a complex network involving many transcription factors and hormones that have a function either in the meristem identity and maintenance or in the initiation and formation of lateral organs.1 The meristem identity of cells in the SAM is correlated with the expression of specific regulatory genes. Class 1 KNOTTED1-like homeobox (KNOX1) transcription factors are expressed in overlapping domains within the SAMs of both monocot and dicot plants, but excluded from the leaf founder cells and primordia.2 KNOX1 genes promote meristem function partly through the repression of biosynthesis of the growth regulator gibberell (GA).3
The YABBY family transcription factors are encoded by a small gene family in Arabidopsis and rice.4,5 All of the Arabidopsis genes show a polar expression pattern and function to determine the abaxial cell fate of one or more above ground lateral organs.4 Activity of the YABBY family of putative transcription factors also contributes to the exclusion of KNOX gene expression from Arabidopsis. Rice YABBY genes are also expressed in leaf primordia and young leaves, but do not show any adaxial/abaxial polar distribution.5,7,8
The meristem stem cell specification requires the activity of the WUSCHEL homeobox gene that is expressed in the meristem organizing center in Arabidopsis.9 The WUS-related homeobox genes (WOX) are suggested to play an important role in region-specific transcription programs early during embryogenesis and lateral organ development in Arabidopsis.10 However, none of the isolated rice or maize WUS orthologues displays an organizing center-type expression pattern in the vegetative SAM as in Arabidopsis,11 while the WOX orthologues from Arabidopsis (PRS) and maize (narrow sheath1/narrow sheath2, NS1/NS2) genes have been shown to function in a lateral domain of the SAM for lateral organ formation.12,13 This suggests that both functional divergence and conservation could be attributed to this class of transcription regulators.
A WOX/YABBY/KNOX/GA Connection
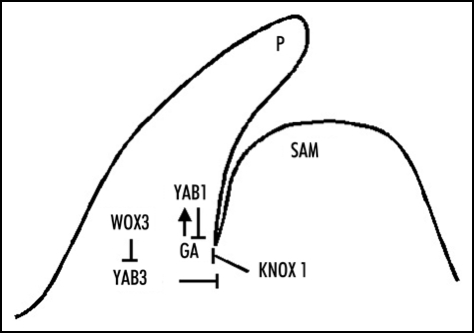
In our recent work, we have established a link between a WOX gene (WOX3) and a YABBY gene (YAB3) in rice.5 Both genes are expressed in the leaf primordia and young leaves but excluded from the SAM. The expression of the genes in young leaves or leaf primordia did not show any adaxial/abaxial polarity. Over-expression of WOX3 repressed the endogenous YAB3 and induced a similar phenotype as YAB3 RNAi plants, which resembles that induced by KNOX1 ectopic expression,14 whereas WOX3 RNAi or YAB3 overexpression produced no visible phenotype. Indeed, rice KNOX1 genes Osh1 and Osh3 are ectopically induced in leaves of WOX3 overexpression or YAB3 RNAi plants, suggesting that YAB3 is required to repress KNOX1 genes in leaves. In addition, WOX3 binds to a conserved DNA element found in YAB3 and is shown to directly repress YAB3, indicating that WOX3 is a transcriptional repressor of YAB3. Thus, the data establish a transcription regulatory hierarchy between WOX3/YAB3/KNOX, which is likely required to regulate the balance between cell division and differentiation during leaf initiation and growth in rice (Fig. 1).
Figure 1.
Functional relationship between WoX3, YAB3, KNoX1 and gibberellin in the control of rice shoot development. SAM, shoot apical meristem. P, leaf primordium.
In a parallel work,8 we have shown that another YABBY gene, YAB1, exhibits a similar expression pattern in the shoot as YAB3 or WOX3. In fact, this expression pattern overlaps with that of key GA biosynthetic genes in rice.15 GA is required to promote cell growth during lateral organ formation from the SAM. YAB1 is induced by GA, but in turn represses the GA biosynthetic gene GA3ox2 by binding to the GA-responsive element (GARE) within the gene, showing that YAB1 is involved in the feedback regulation of GA biosynthesis in the shoot. Accordingly, overexpression of YAB1 induced a semi-dwarf phenotype that could be complemented by applied GA. These data establish that YAB1 is involved in the control of GA homeostasis during leaf cell growth, while previous work has shown that KNOX1 genes exclude GA biosynthesis from the SAM to maintain the SAM cell identity.16 Therefore, GA biosynthesis turns out to be a common target of two otherwise antagonistic transcriptional regulators.
Conclusions and Perspectives
Therefore, there are two parallel pathways involving YABBY genes to control cell division and differentiation during rice leaf growth (Fig. 1). Studying the interplay between transcriptional regulators and growth hormones is important to decipher the mechanism controlling mersitem activity and organogenesis. Further work is required to show if there is any link between the two pathways in the control of rice leaf development. Importantly, the relationship between GA and the other YABBY genes needs to be determined. In addition, it remains to know whether other YABBY members (including YAB3) also bind to the GARE, as rice KNOX1 genes likely contain this DNA-binding motif. The repression of KNOX1 genes in leaves also involves the myb transcription factor AS1/RS2/PHAN,17 which likely induces chromatin modification to maintain KONX1 gene repression through interaction with histone chaperone proteins.18 Therefore, experiments will be required to show whether the YAB3 is directly involved in the KNOX1 repression and whether the transcriptional repression mediated by YABBY proteins and WOX3 involves also chromatin modification.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4279
References
- 1.Shani E, Yanai O, Ori N. The role of hormones in shoot apical meristem function. Curr Opin Plant Biol. 2006;9:484–489. doi: 10.1016/j.pbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Reiser L, Sanchez-Baracaldo P, Hake S. Konts in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol. 2000;42:151–166. [PubMed] [Google Scholar]
- 3.Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol. 2002;12:1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- 4.Bowman JL. The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol. 2000;3:17–22. doi: 10.1016/s1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 5.Dai M, Hu Y, Zhao Y, Zhou DX. A wuschel-like homeobox gene represses a Yabby gene expression required for rice leaf development. Plant Physiol. 2007;144:121–123. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumaran MK, Bowman JL, Sundaresan V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell. 2002;14:2761–2770. doi: 10.1105/tpc.004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX. The Rice YAB1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007 doi: 10.1104/pp.107.096586. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 10.Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- 11.Nardman J, Werr W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol Biol Evol. 2006;12:2492–2504. doi: 10.1093/molbev/msl125. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001;15:3355–3364. doi: 10.1101/gad.931001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nardmann J, Ji J, Werr W, Scanlon MJ. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development. 2004;131:2827–2839. doi: 10.1242/dev.01164. [DOI] [PubMed] [Google Scholar]
- 14.Sentoku N, Sato Y, Matsuoka M. Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev Biol. 2000;220:358–364. doi: 10.1006/dbio.2000.9624. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 2003;35:104–115. doi: 10.1046/j.1365-313x.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 2001;15:581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 18.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MCP. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 Interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]