Abstract
To better understand plant vacuolar functions and identify new transporters present on the tonoplast, a proteomic work was initiated on Arabidopsis thaliana. A procedure was developed to prepare highly purified vacuoles from protoplasts isolated from Arabidopsis cell cultures, and a proteomics approach was designed to identify the protein components present in both the membrane and soluble fractions of the vacuoles. This procedure allowed the identification of 650 proteins, 2/3 of which copurify with the hydrophobic membrane fraction and 1/3 with the soluble fraction. With regard to function, only 20% of the proteins identified were previously known to be associated with vacuolar activities.
Key WordS: Arabidopsis, vacuole, proteome, tonoplaste, transporter
Membranes, and proteins they contain, occupy a fundamental place in the complex organization of the cell because of their position at the interface between cells or between cell compartments. They control exchange of signals and solutes and allow the compartmentalization of specific biochemical pathways in subcellular organelles. Plant cell vacuoles are surrounded by the tonoplast membrane and play a fundamental role in plant cells: storage of metabolites, maintenance of turgor pressure, digestion of cytoplasmic constituents, pH regulation and ion homeostasis.1 Surprisingly, in spite of the dominant role of the vacuolar compartment very few transporters have been identified so far. Recently, proteomic analyses of the tonoplast have been published.2–4 Shimaoka et al.2 identified a large number of mostly soluble proteins within their vacuolar fractions. The most complete study published identified 402 proteins.4 However, only 29 were putative or known transporters. Taken together, all these studies indicated the need to extend the knowledge of the vacuolar proteome (vacuome).
Given the tremendous dynamic range of protein abundances and the extraordinary complexity of proteomes, enrichment of low abundance proteins is one of the key issues we focus on. To that aim, we have developed “divide-and-conquer” strategies that combine, protein and/or organelle separation to study specific subsets of the proteome comprehensively followed by integration of multidimensional fractionation/separations prior to MS analysis. Taking into account the fact that vacuoles play an essential role in bulk protein turnover and in the autophagic engulfment of cytoplasm and organelles, it could be difficult to discriminate between the genuine organelle resident proteins and contaminants. To characterize our preparation as completely as possible, potential cross contaminations were estimated by western blots and specific activity analyses (α-mannosidase being used as vacuolar marker). The enrichment factor was estimated to be around 42-fold when compared to the specific activity of the same marker in protoplast extract with an average yield of 2.1%.
We first undertook the identification of the most abundant total vacuolar proteins. To that end, the most intense bands of vacuole extract, separated by SDS-PAGE, were digested and peptide mixtures were submitted to LC-MS/MS. Surprisingly, among the most abundant proteins which are most by known vacuolar proteins, the SOUL heme-binding protein was identified as a major component. SOUL proteins are heme-binding proteins identified in mammalian cells and may be involved in heme transfer or binding of free heme to prevent damage by reactive oxygen species.5,6 The vacuole could act as both a storage and a buffer compartment, enhancing iron supply in high demand situations, in addition to detoxifying when excesses occur.
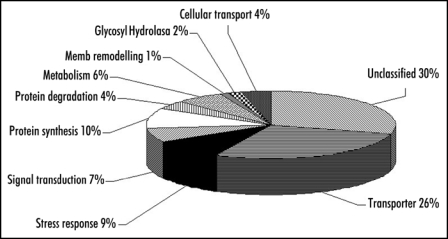
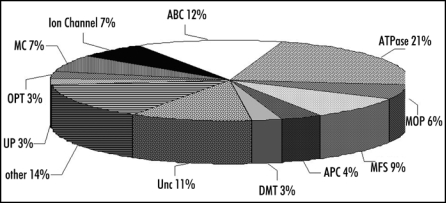
To assess our sample quality and identify the major tonoplastic proteins, vacuoles were first frozen and thawed, the membrane fraction was washed and the most hydrophobic fraction was then analyzed. To quickly qualify and estimate the purity of the membrane fraction, an in-solution digestion was chosen. It has been established that the real intrinsic membrane proteins are both hydrophobic and proteolytically resistant proteins. Thus, to improve the trypsin enzyme hydrolysis, additives such as surfactants, are commonly used to improve protein solubility and hence facilitate a more complete hydrolysis. The drawback of this approach is that denaturants often reduce the proteolytic activity of enzymes and interfere with MS and LC. At this step, an atypical proteolysis in a mixed-organic-aqueous solvent system was performed to digest the protein mixture and to allow identification of the most abundant hydrophobic proteins. The resulting peptide mixture was analyzed and 122 proteins were identified from a very low amount of total proteins (2 × 0.15 µg). No obvious contaminants from mitochondria, chloroplast or other membrane systems were identified. However, a few cytosolic components were identified in the mixture. Thus, we concluded that the hydrophobic starting material was sufficiently pure for further fractionation and MS analysis to obtain meaningful proteome data. Using both in-gel and in-solution digestion protocols, 416 non-redundant proteins were identified from the membrane fraction. One hundred ninety-five were integral membrane proteins, five had transmembrane beta barrel, 29 did not have any transmembrane domains but were known to be part of membrane complexes (H+-ATPase), and 31 were predicted to have covalent lipid modifications leading to their insertion into the membrane. The proteins are categorized into ten classes (Fig. 1). Among these, transporters (110 identified) represent 26% of the proteins. Across the panel of transporters (Fig. 2), the main representative superfamily was the ABC transporters (14 identifications) belonging to five different subfamilies.7 Ten identified as MRP1-8, MRP10 and MRP14 are members of the Multidrug Resistance associated Protein family. However, the degree of identities shared between members of this protein family ranges from 33% to 87%8 making their unambiguous identification difficult without a high protein coverage. Here, we want to highlight the problem of ambiguous MS-based identification, often ignored in the literature. In quite a few instances, it is possible that the same MS/MS spectrum matches more than one protein. In many other cases, only one peptide sequence is interpreted from the MS/MS spectrum, but it can match more than one protein, leading to misidentification of proteins. We would like to point out that our “divide-and-conquer” strategy allowed us to reach a level of sequence coverage which lowers number of possible ambiguities. Based on the protein coverage obtained using the mixed-organic-aqueous digestion, MRP10 seems to be the most abundant ABC transporter in cultured Arabidopsis cells. So far, only AtMRP1, AtMRP2 and AtMRP4 have been localized to the tonoplast by classical methods.9–11 Recently, Dunkley et al have assigned transporters to specific organelles by LOPIT technology,11 and shown that the tonoplast is dominated by proteins involved in trans-membrane transport, including eight ABC transporters (MRP1-6, MRP10 and TAP2).
Figure 1.
Classification of the tonoplastic proteins identified according to Gene ontology function entries displayed as pie diagrams. Proteins were categorized into ten major groups: transporters, stress response, signal transduction, metabolism, cellular transport, protein synthesis and degradation, glycosyl hydrolase, membrane remodelling and unclassified function.
Figure 2.
Overview of the transporter families identified in the tonoplast. The transporters were classified according to Transport DB (www.membranetransport.org): ABC, ATP-Binding Cassette Transporter; MFS, Major Facilitator Superfamily; MOP, Multidrug/Oligosaccharidyl-lipid/Polysaccharide; DMT, Drug/Metabolite Transporter; OPT, Oligopeptide Transporter; KuP, K+ Uptake Permease; APC, Amino Acid-Polyamine-Organocation; MC, Mitochondrial Carrier; ATPase; Unc, unclassified.
A few unexpected proteins, such as TolB or Niemann-Pick C1 protein (NPC1) were identified in the tonoplast fraction. For exemple, TolB, a periplasmic protein found in Gram-negative bacteria, is one of the Tol proteins involved in the translocation of group A colicins.12 The exact role of the Tol system remains to be determined. Its presence in the tonoplast is probably linked to membrane biogenesis, as has been suggested for the Gram-negative organisms. NPC1 protein is a glycoprotein that was shown to reside primarily in mammalian late endosomes. A number of observations suggest that NPC1 may be related to prokaryotic efflux pumps and thus it may act as a molecular pump,13 a cholesterol transporter14 or be involved in docking/fusion events.15 From our results, we highlighted another important uncharacterized membrane proteins family: the band 7 protein family (ten members out of the twenty predicted from the Arabidopsis thaliana genome were identified in the tonoplastic fraction). These SPFH-domain proteins16 which have hydrophobic stretches but no classical transmembrane domain, are of prime interest because they are putatively involved in cell cycle and ion channel control17,18 or may be actors for the recruitment of multiprotein complex17 formation with membrane-associated proteases19,20 putatively involved in biogenesis, maintenance and turn over of this organelle.
The use of subcellular fractionation permits a simplification of the proteome and provides a practical step towards the ultimate description of the entire cell proteome. The quality of the purified vacuolar fraction, as assessed by enzymatic and immunological assays, was the first criterion for establishing the likelihood of vacuolar localization of the proteins identified in this study. To further characterize the system, the sub-cellular localization of several proteins was confirmed by transient expression in transfected tobacco plants and in Arabidopsis protoplasts of GFP-fusion proteins. As expected, and taking into account the overall results presented previously, the vacuolar localization of all the GFP tagged proteins was confirmed.
Through our proteomic strategy, it seems clear that the tonoplast is a complex structure that receives membrane and protein contributions from a variety of subcellular sources and pathways. The high degree of protein diversity of the tonoplast membrane is indicative of a highly complex organelle. Further validation of the results presented here by relevant functional studies should provide a better explanation for the biogenesis and maintenance of these unique organelles.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4415
References
- 1.De DN. Plant Cell Vacuoles: An Introduction. Australia, Collingwood: CSIRO publishing; 2000. pp. 79–114. [Google Scholar]
- 2.Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T. Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol. 2004;45:672–683. doi: 10.1093/pcp/pch099. [DOI] [PubMed] [Google Scholar]
- 3.Szponarski W, Sommerer N, Boyer JC, Rossignol M, Gibrat R. Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics. 2004;4:397–406. doi: 10.1002/pmic.200300607. [DOI] [PubMed] [Google Scholar]
- 4.Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T. Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J Biol Chem. 1998;273:31388–31394. doi: 10.1074/jbc.273.47.31388. [DOI] [PubMed] [Google Scholar]
- 6.Sato E, Sagami I, Uchida T, Sato A, Kitagawa T, Igarashi J, Shimizu T. SOUL in mouse eyes is a new hexameric heme-binding protein with characteristic optical absorption, resonance Raman spectral, and heme-binding properties. Biochemistry. 2004;43:14189–14198. doi: 10.1021/bi048742i. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez Fernandez R, Davies TGE, Coleman JOD, Rea PA. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 8.Kolukisaoglu HU, Bovet L, Klein M, Eggmann T, Geisler M, Wanke D, Martinoia E, Schulz B. Family business: The multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta. 2002;216:107–119. doi: 10.1007/s00425-002-0890-6. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Sanchez-Fernandez R, Li ZS, Rea PA. Enhanced multispecificity of arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J Biol Chem. 2001;276:8648–8656. doi: 10.1074/jbc.M009690200. [DOI] [PubMed] [Google Scholar]
- 10.Geisler M, Girin M, Brandt S, Vincenzetti V, Plaza S, Paris N, Kobae Y, Maeshima M, Billion K, Kolukisaoglu UH, Schulz B, Martinoia E. Arabidopsis immunophilin-like TWD1 functionally interacts with vacuolar ABC transporters. Mol Biol Cell. 2004;15:3393–3405. doi: 10.1091/mbc.E03-11-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, Watson RB, Dupree P, Lilley KS. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazzaroni JC, Dubuisson JF, Vianney A. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie. 2002;84:391–397. doi: 10.1016/s0300-9084(02)01419-0. [DOI] [PubMed] [Google Scholar]
- 13.Davies JP, Chen FW, Ioannou YA. Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science. 2000;290:2295–2298. doi: 10.1126/science.290.5500.2295. [DOI] [PubMed] [Google Scholar]
- 14.Amigo L, Mendoza H, Castro J, Quinones V, Miquel JF, Zanlungo S. Relevance of Niemann-Pick type C1 protein expression in controlling plasma cholesterol and biliary lipid secretion in mice. Hepatology. 2002;36:819–828. doi: 10.1053/jhep.2002.35617. [DOI] [PubMed] [Google Scholar]
- 15.Ioannou YA. Guilty until proven innocent: The case of NPC1 and cholesterol. Trends Biochem Sci. 2005;30:498–505. doi: 10.1016/j.tibs.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: Implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Arnadottir J, Keller C, Caldwell GA, Yao CA, Chalfie M. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JZ, Hayashi H, Ebina Y, Prohaska R, Ismail-Beigi F. Association of stomatin (band 7.2b) with Glut1 glucose transporter. Arch Biochem Biophys. 1999;372:173–178. doi: 10.1006/abbi.1999.1489. [DOI] [PubMed] [Google Scholar]
- 19.Kihara A, Akiyama Y, Ito K. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 1996;15:6122–6131. [PMC free article] [PubMed] [Google Scholar]
- 20.Green JB, Fricke B, Chetty MC, von During M, Preston GF, Stewart GW. Eukaryotic and prokaryotic stomatins: The proteolytic link. Blood Cells Mol Dis. 2004;32:411–422. doi: 10.1016/j.bcmd.2004.01.016. [DOI] [PubMed] [Google Scholar]